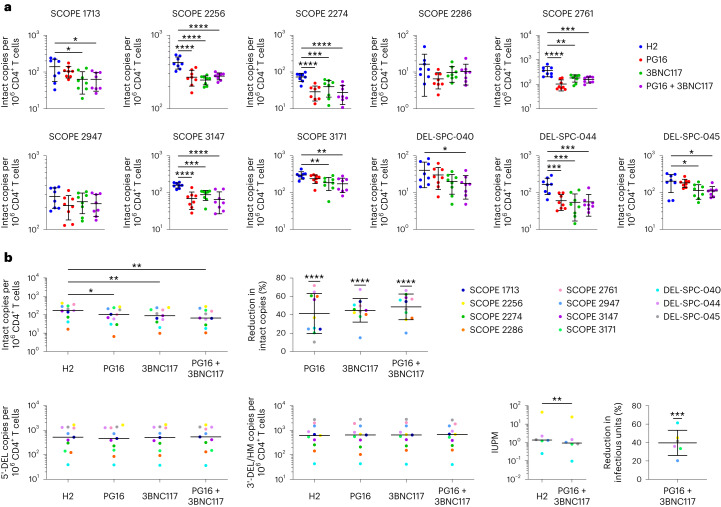
Fig. 5. PG16-Db and 3BNC117-Db eliminate cells harboring intact and inducible proviruses.
a, Frequency of intact provirus+CD4+ T cells after treatment with PMA + I and coculture with autologous NK cells in the presence of H2-Db, PG16-Db, 3BNC117-Db or PG16-Db + 3BNC117-Db. Data are reported as intact HIV-1 copies per million viable CD4+ T cells (mean, s.d.) from eight replicates. The significance was determined by one-way ANOVA followed by Dunnett’s test for multiple comparisons: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. b, Average frequencies of intact provirus+ CD4+ T cells (top left), 5′-deleted provirus+ CD4+ T cells (bottom far left) and 3′-deleted/hypermutated provirus+CD4+ T cells (bottom middle left) after treatment with PMA + I and coculture with autologous NK cells as in a (mean, s.d.) across all 11 participants assessed. The significance was determined by one-way ANOVA followed by Dunnett’s test for multiple comparisons: *P < 0.05, **P < 0.01. Average IUPM viable CD4+ T cells (bottom middle right) after treatment with PMA + I and coculture with autologous NK cells as in a (mean, s.d.) across six participants assessed. The significance was determined by paired, two-sided Student’s t-test: **P < 0.01. Average frequency of intact provirus+ CD4+ T cells (top middle) or average IUPM viable CD4+ T cells (bottom far right) relative to H2-Db. Data reported as a percentage reduction (mean, s.d.) from eleven or six participants, respectively. The significance was determined by Student’s t-tests against a theoretical mean of 0% reductions: ***P < 0.001, ****P < 0.0001.