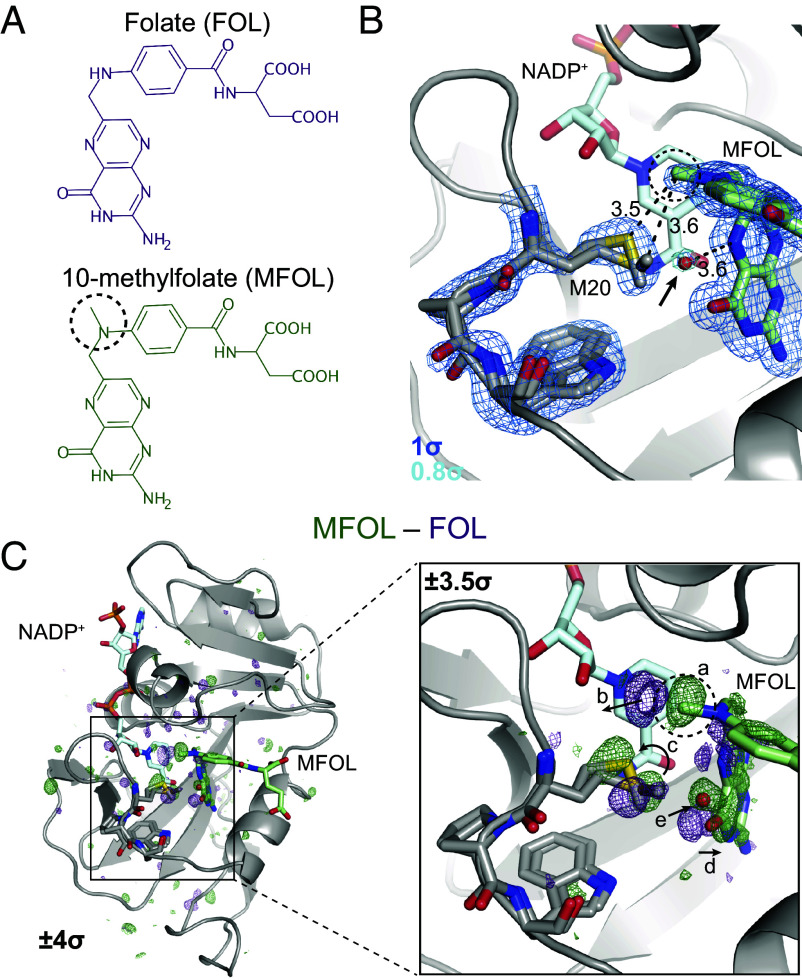
Fig. 2.
Ligand-dependent conformational changes illustrate Met20 solvent gating. (A) Chemical structures of FOL and MFOL. (B) Refined structure and electron density map of the ecDHFR:NADP+:MFOL complex. The 10-methyl group is in close contact with the Met20 sidechain, and a water (red sphere; indicated by an arrow) can be resolved within 3.6 Å of the N5 nitrogen of MFOL. The map is contoured at (blue mesh; carved within 1.5 Å of shown atoms) and (light blue mesh; carved within 1.5 Å of shown water). (C) isomorphous difference map, phased with the MFOL-bound model. The overview shows the difference electron density induced by the 10-methyl substituent (), and the Inset highlights the structural differences observed in the active site (, carved within 3.0 Å of shown atoms). The added methyl group (label a) displaces an ordered water (label b), alters the rotamer distribution of Met20 (label c), rotates the pterin ring (label d), and leads to the introduction of an ordered water near the N5 nitrogen (label e). The 10-methyl substituent is indicated with a dashed circle in each panel.