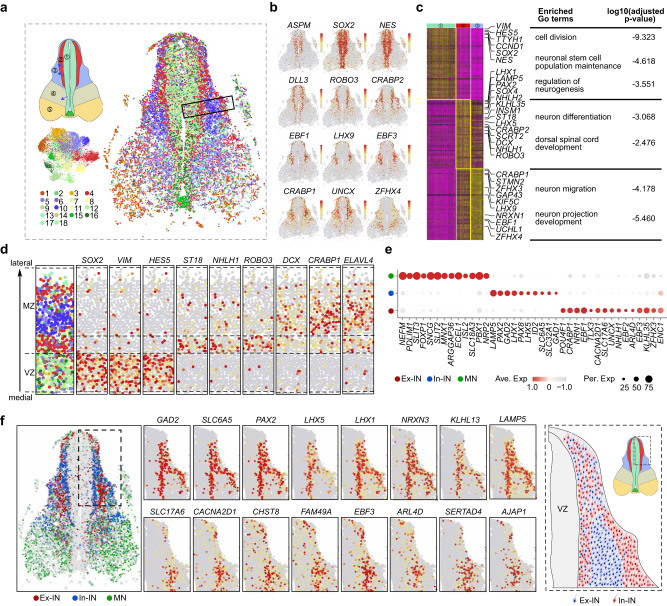
Fig. 3. Spatially resolved characterization of the mediolateral organization pattern in the developing human spinal cord.
a Unsupervised clustering in TF-seqFISH dataset identified 18 molecularly defined clusters and their corresponding spatial localization in a slice of developing human spinal cord at GW8. Individual cells are represented by dots and colored based on the clusters, revealing five distinct anatomical regions based on spatially restricted cell distribution. To aid clarity, a schematic illustration is also provided, where distinct regions are denoted by digits and blue arrows indicate migratory directions of neurons in the spinal cord. b Visualization of the imputed gene expression profiles in the developing human spinal cord, with each dot representing an individual cell and color coded according to the expression level (red, high; gray, low). c A heatmap depicting the expression profiles of DEGs among cells located in Regions 1, 2, and 3 of the developing human spinal cord is presented, with enriched GO terms for the DEGs listed. d Visualization of laminar cell organization and gene expression, including TFs and non-TF genes, along the mediolateral axis in the dorsal horn of the developing human spinal cord facilitated by utilizing the imputed data. Each dot represents an individual cell and is colored based on expression level (red, high; gray, low). The leftmost column is a zoomed-in image of the boxed area in a. e A dot plot illustrating the expression profiles of DEGs among Ex-INs, In-INs, and MNs in the developing human spinal cord. f The spatial organization of Ex-INs, In-INs, and MNs in the developing human spinal cord is displayed, with the spatially-restricted expression of genes specific for Ex-INs and In-INs in the dorsal horn also depicted in the boxed area based on the imputed data. A schematic representation of the sandwich-like arrangement of Ex-INs and In-INs in the dorsal horn is provided for clarity.