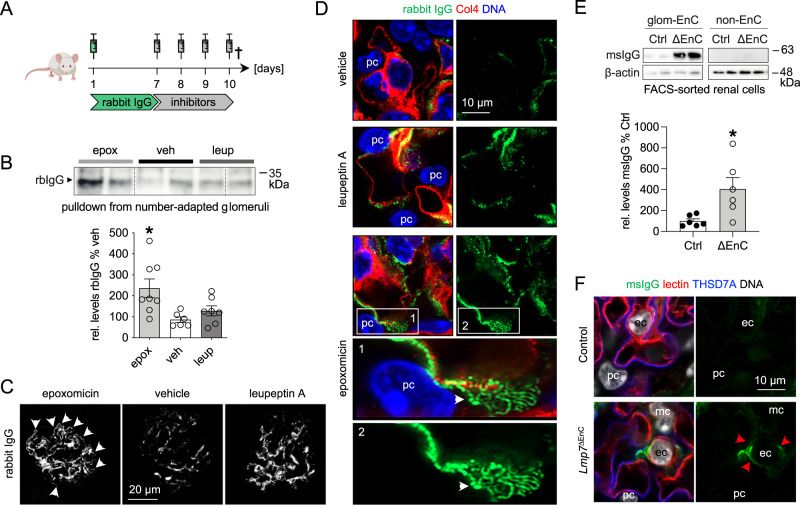
Fig. 5. Proteasome impairment results in glomerular IgG accumulation.
A Scheme depicting the experimental setup. Mice were treated with the proteasome inhibitor epoxomicin (epox, 0.5 µg/g bodyweight), the lysosomal inhibitor leupeptin A (leup, 40 µg/g bodyweight) or equal volumes of DMSO (veh, vehicle, 125 µl) on four consecutive days following an initial injection with 160 µl of unspecific rabbit IgG (rbIgG) or PBS. Mice were perfused and kidneys were removed on day 10. Micrographs were analyzed from 3 individual experiments, with 3 micrographs per group. B Immunoblot quantification of deposited rbIgG following rbIgG-pulldown from isolated glomeruli. Graph exhibits densitometric analysis, mean ± SEM, n = 6 (veh), n = 7 (leup), n = 8 (epox), pooled data from 2 independent experiments, *p = 0.013, one-way ANOVA with Bonferroni’s post-test for multiple comparisons. C Confocal micrographs depicting rbIgG (white) deposition pattern in glomeruli on day 10, note the linear deposition at the GFB (white arrows) of the epoxomicin-treated mouse. D High-resolution confocal micrographs resolving rabbit IgG (green) accumulation pattern in epoxomicin-treated mice at the GFB. White arrows point toward linear meandering rbIgG accumulations in the subepithelial space. The glomerular basement membrane is marked by collagen type 4 (red) and DNA is depicted in blue (Hoechst). E Lmp7ΔEnC and control littermates were analyzed after induction of β5i knockout. Following kidney perfusion with PBS, glomerular (GEnCs) and non-endothelial kidney cells were FACS-sorted. Immunoblot quantification of msIgG, densitometric analysis relative to control littermate cell populations, mean ± SEM, n = 6 mice of 2 pooled independent experiments, *p = 0.0152, two-sided Mann–Whitney U. Equal loading was ensured by loading equal numbers of FACS-sorted endothelial cell populations between genotypes. F Experimental kidneys of Lmp7ΔEnC and controls were additionally perfused with AF647-Lycopersicon esculentum lectin (here: red) to demarcate the endothelial glycocalyx. Following zinc-fixation, sections were stained for msIgG (green) and the podocyte (pc) foot process protein THSD7A to enable localization of msIgG, DNA (white, Hoechst). Red arrows point toward msIgG accumulation underneath the glycocalyx within GEnCs (ec) in the Lmp7ΔEnC mouse, mc mesangial cell. Scheme was created with BioRender.com. Source data are provided as a Source Data file.