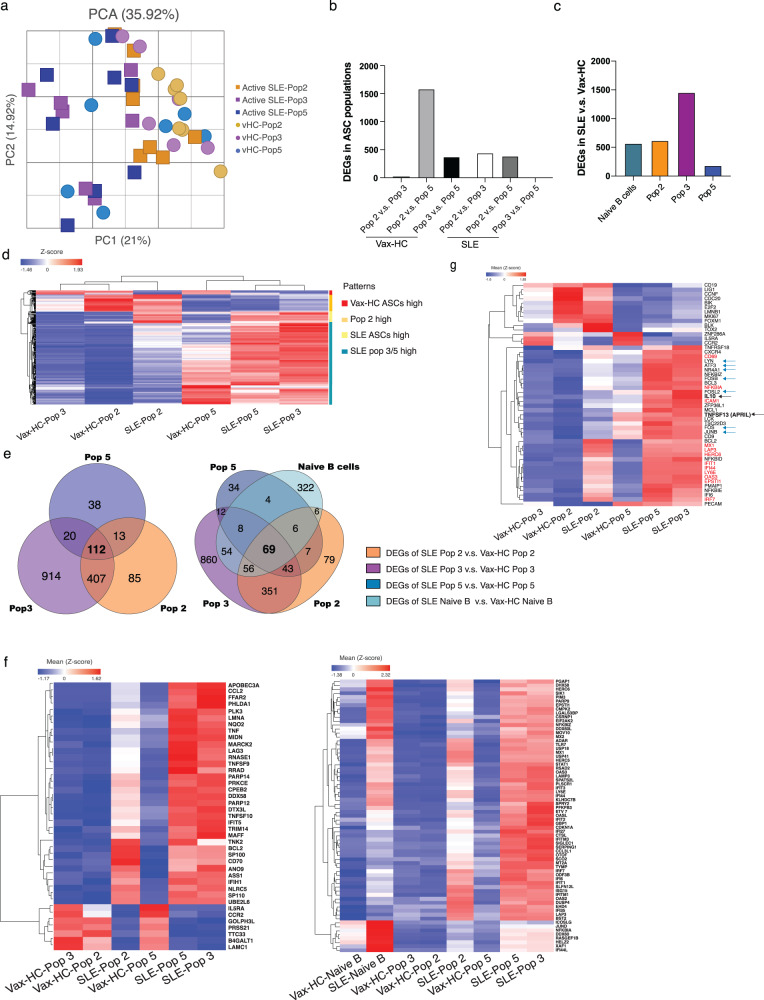
Fig. 6. SLE ASC express a distinct transcriptome.
Peripheral ASC populations from 9 active SLE patients and 7 healthy subjects post influenza vaccination were sorted and their transcriptomes analyzed by RNA sequencing. a Principal component analysis (PCA) of differentially expressed genes (DEGs) (at least two-fold change of expression and FDR < 0.05) detected from all pairwise comparisons of the six groups. b DEGs between ASC pops within SLE and Vax-HC. Bar plots show DEGs within populations from same disease groups, including both active SLE patients and heathy subjects post vaccination, respectively. c Bar plots show DEGs between SLE and post-vax HC for each ASC population and naive B cells. d Heatmap of z-score normalized RPKM expression for all 2200 DEGs identified, and their distribution across four clusters indicated by the color code of the Y axis to the right: High expression in Vax-HC ASC; high expression restricted to pop2 whether SLE or HC; high expression in all SLE ASC; and high expression restricted to SLE pops 3 and 5. e Venn diagrams show DEG sharing between all ASC Pops identified from the comparison of SLE and vaccinated HC (left), and overlap of DEGs among all ASC Pops and naive B cells by comparing SLE and vaccinated HC (right). f Heatmap of z-score normalized RPKM expression for 43 overlapping DEGs shared between all ASC populations but not by naive B cells (left heatmap), and for the 69 genes shared between ASC populations and naive B (right heatmap). g Heatmap of z-score normalized RPKM expression for top DEGs of immunological relevance. IFN-stimulated genes are colored in red. Overexpression of IL-10 and APRIL is indicated in bold and black arrows. Transcription factors (TF) known to determine a large fraction of the SLE naive B cell transcriptome through epigenetic regulation are indicated by blue arrows.