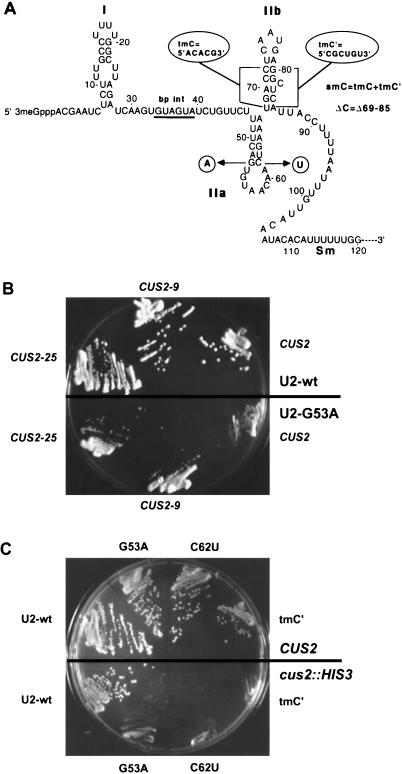
FIG. 1.
(A) Secondary structure model of the 5′ end of yeast U2 snRNA. Mutations G53A, C62U, A61Δ, tmC (68ACACG72), tmC′ (80CGCUGU85), smC (tmC plus tmC′), ΔC (deletion of nucleotides 69 to 85), and SMB (58AAC60 plus 99UUG101) discussed in the text are indicated. (B) Suppression of G53A and C62U by different CUS2 alleles. Strain BJ81 was transformed with either wild-type (wt) U2 (top half of plate) or the cold-sensitive G53A mutation (bottom half) and the indicated CUS2 allele on a plasmid. Strains were streaked onto glucose-containing medium and incubated at 18°C for 5 days. (C) Enhancement of U2 mutations by the cus2::HIS3 disruption. Strain D2 (top half of plate) or D2CUS2KO (bottom half) was transformed with a plasmid carrying the indicated U2 allele and streaked on 5-fluoroorotic acid at 26°C to shuffle out the wild-type U2 gene on pCH1122. Growth was for 4 days.