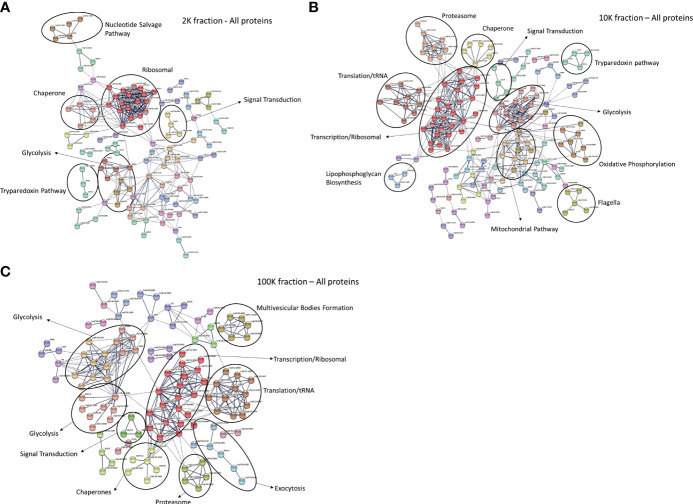
Figure 2.
Protein-protein interaction networks of total proteins detected in fractions of Leishmania major-derived extracellular vesicles display notable variations. Networks were generated using total proteins detected by LC-MS/MS in (A) 2K, (B) 10K, and (C) 100K fractions of Leishmania major-derived EVs. In the 2K/apoptotic body fraction (A), notable clusters include the Nucleotide Salvage Pathway, Ribosomal, Chaperone, Signal transduction, Glycolysis and the Tryparedoxin pathway. In the 10K/Ectosome fraction (B), clustering patterns include Proteasome, Chaperone, Tryparedoxin pathway, Translation/tRNA, Ribosome, Signal transduction, Glycolysis, Lipophosphoglycan biosynthesis, Mitochondria, Oxidative phosphorylation, and Flagella. In the 100K/exosome fraction (C) clusters include Glycolysis, Signal transduction, Chaperone, Proteasome, Translation/tRNA, Ribosome, along with unique clusters representing Multivesicular Body Formation and Exocytosis. Protein-protein interaction networks of the identified proteins were created using String database (https://www.string-db.org) using the highest confidence of interaction (>0.900), showing only connected proteins in the network and using an MCL inflation parameter of 3.