Figure 2.
RNA sequencing analysis revealed the acquisition of a fetal/regenerative/revival signature in 2D HC
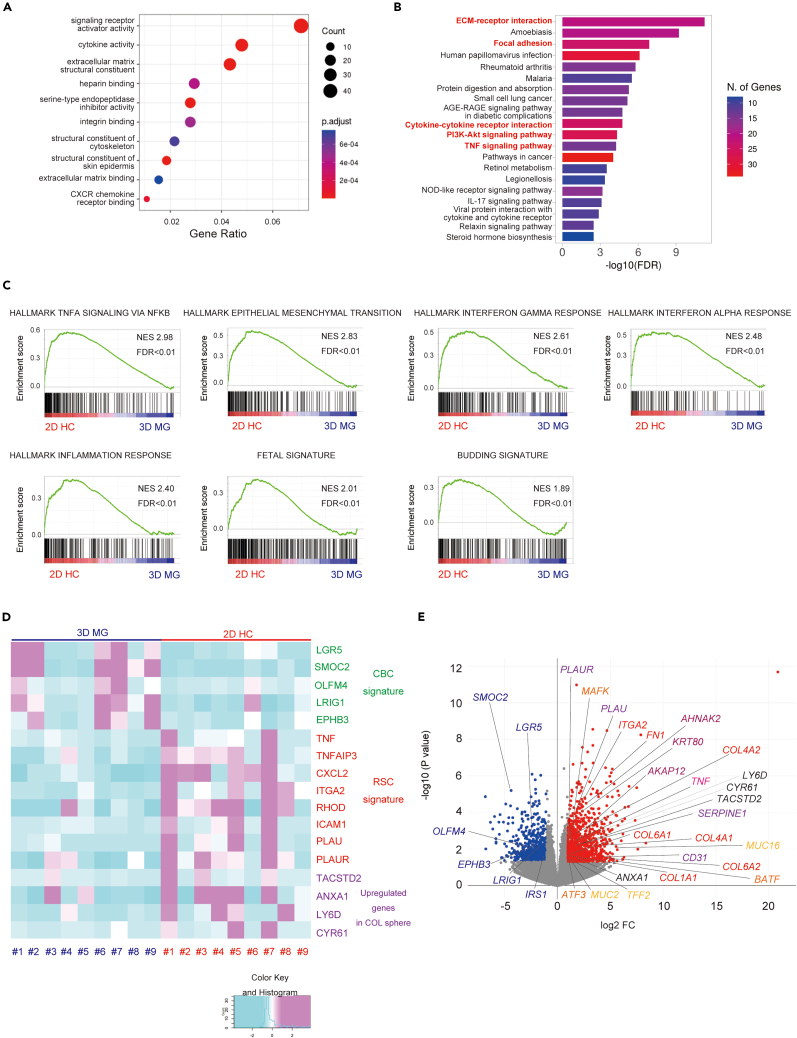
(A) The top 10 GO-TERM (molecular function) of the upregulated gene set in 2D HC compared to 3D MG (2D HC gene set) is shown.
(B) The top 20 KEGG pathways enriched in the 2D HC gene set are shown.
(C) The GSEA of hallmark gene sets, fetal gene signature, and tumor budding signature enriched in 2D HC gene set are shown. NES; normalized enrichment score, FDR; the false discovery rate.
(D) The heatmap displays Z score-transformed relative expression levels of genes for crypt base columnar (CBC) signature (LGR5, SMOC2, OLFM4, LRIG1 and EPHB3), regenerative stem cell (RSC) signature (TNF, TNFAPI3, CXCL2, ITGA2, RHOD, ICAM1, PLAU and PLAUR), and representative upregulated genes in normal COL sphere (TACSTD2, ANXA1, LY6D and CYR61) in nine cultures of 3D MG and 2D HC. COL; collagen.
(E) A volcano plot of differentially expressed genes included in 3D MG and 2D HC gene sets is indicated. Representative genes are indicated in a plot. See also Table S1.