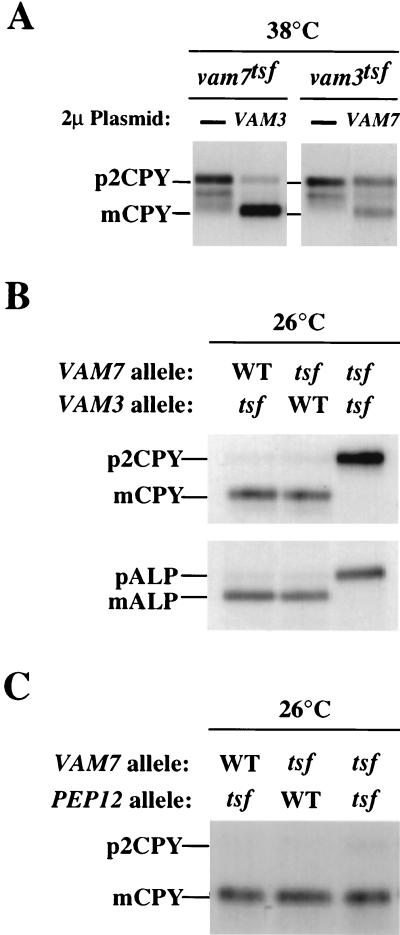
FIG. 5.
Genetic interactions between VAM7 and VAM3. (A) Suppression of vam7tsf and vam3tsf mutant cells. TKSY43 (vam7Δ) or TDY2 (vam3Δ) containing tsf alleles of either vam7 (pTKS43ts-167) or vam3 (pVAM3.6-416), respectively, were transformed with a multicopy vector (2μm) containing no insert, VAM7 (pTKS23), or VAM3 (pVAM3.426) as indicated. Cells grown at 26°C were harvested, incubated at the permissive or nonpermissive temperature for 10 min, and labeled with [35S]cysteine and [35S]methionine for 10 min. After the labeling, cells were chased with unlabeled cysteine and methionine for 30 min and harvested by TCA precipitation. Lysates from these cells were immunoprecipitated with the indicated antibodies and analyzed as previously described. (B) Synthetic interactions between vam7tsf and vam3tsf. Double-mutant strains (vam3Δ vam7Δ) harboring the vam3tsf allele (pTKS30 and pVAM3.6-416), the vam7tsf allele (pTKS43ts-167 and pVAM3-416), or both tsf alleles (pTK43ts-167 and pVAM3.6-416) were incubated at 26°C for 10 min and labeled with [35S]cysteine and [35S]methionine for an additional 10 min. After the labeling, the cells were chased with unlabeled cysteine and methionine for 45 min, harvested, and assayed for vacuolar protein sorting as described above. (C) Double-mutant cells (pep12-60 vam7Δ) were transformed with CEN plasmids containing VAM7 (pTKS30) or the vam7tsf allele (pTKS43ts-167). Single- and double-mutant strains were assayed for CPY sorting as described above.