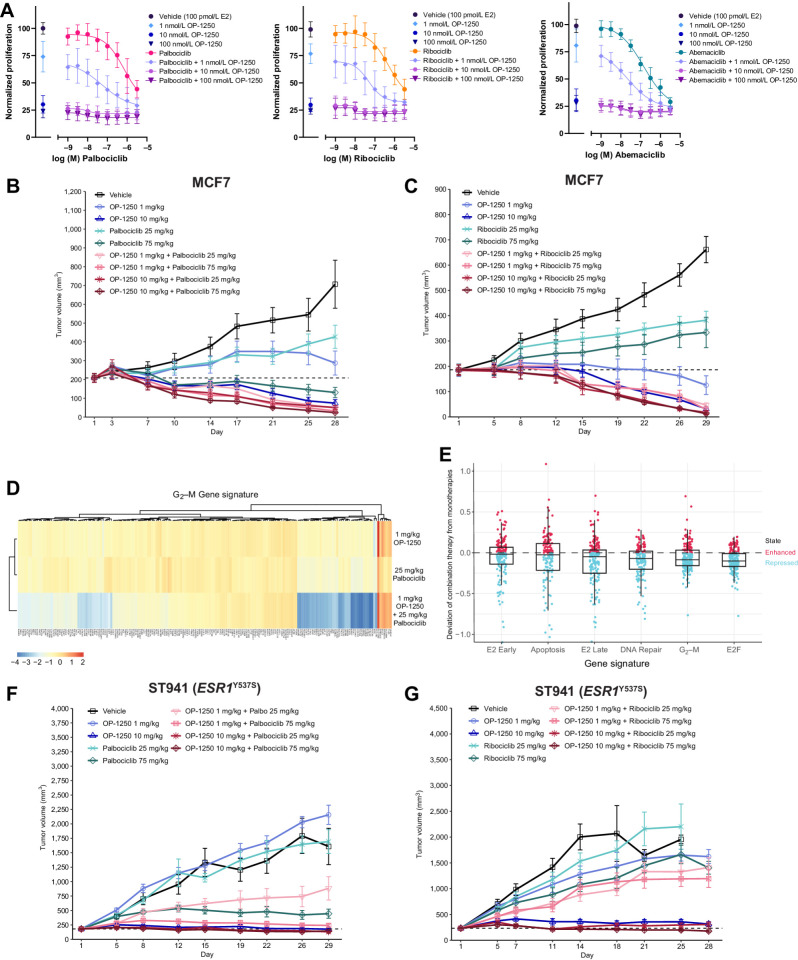
Figure 5.
Addition of OP-1250 improves efficacy of CDK4/6 inhibitors in vitro and in vivo. A, Cellular proliferation assessed by CyQUANT reagent of MCF7 cells treated for 7 days with the CDK4/6 inhibitors palbociclib, ribociclib, or abemaciclib alone or in combination with OP-1250. Dose of indicated CDK4/6 inhibitor is shown on the x-axis, with samples not treated with CDK4/6 inhibitor shown to the left of the axis break. Proliferation is normalized to 100 pmol/L E2 vehicle and shown as mean and SD across three experiments. B and C, Mean and SEM tumor volume over time with monotherapy and combination treatments in the MCF7 breast cancer model, with dotted line representing tumor stasis. D, Heat map showing expression of genes associated with the G2–M gene signature of n = 4 MCF7 xenograft tumors from OP-1250 and palbociclib low dose monotherapy and combination therapy groups. Red corresponds to high expression relative to vehicle and blue corresponds to low expression. E, Differential gene expression comparison between combination therapy and monotherapy MCF7 xenograft samples in hallmark gene sets. Each datapoint represents a gene in the listed gene signature. Vertical axis depicts a datapoint's distance to the y = x diagonal, where 0 represents no change from expected result based on monotherapy values. F and G, Mean and SEM tumor volume over dosing interval with monotherapy and combination treatments in the ESR1Y537S ST941 PDX model, with dotted line representing tumor stasis.