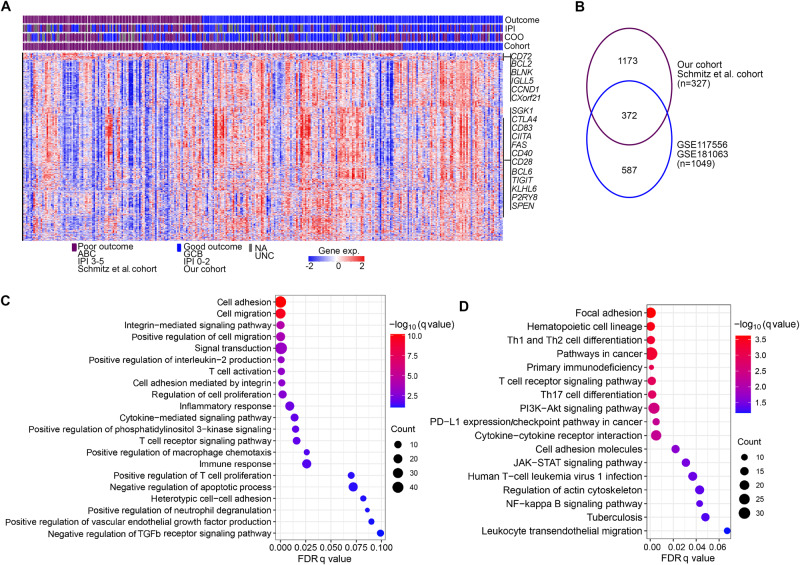
Fig. 3. Gene expression pattern in tumors derived from DLBCL patients with poor and good outcomes at two years following R-CHOP treatment.
The analysis of differentially expressed genes (DEGs) was conducted separately in the RNAseq dataset (our cohort/Schmitz et al. cohort, n = 327) and the microarray dataset (GSE117556/GSE181063; n = 1049). A threshold of FDR q < 0.1 and fold change >1.2 was used to define DEGs from each dataset. A The heatmap illustrating DEGs between the two outcome groups in the RNAseq dataset. Each column represents a sample, ordered by outcome groups. Within each group, samples were ordered following the input of the data accordingly. Each row represents a gene. Selected genes that are frequently mutated in B-cell lymphomas or important for immune responses are highlighted in the figure. B The numbers of overlapping DEGs between the RNAseq dataset and the microarray dataset. GSEA analysis of the overlapping DEGs from (B) (C: Gene Ontology biological pathway, (D): Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway).