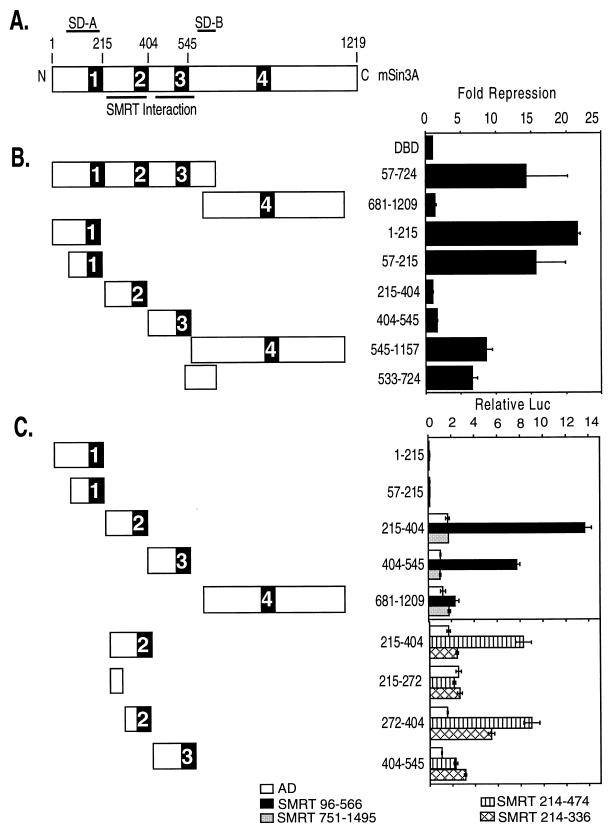
FIG. 3.
Comparative mapping of repression and SMRT interaction domains located within mSin3A. (A) Schematic representation of mSin3A. The locations of four putative paired amphipathic helical domains, PAH1 through -4, are shown. Also depicted are the locations of two domains within mSin3A (SD-A and SD-B) that are able to repress transcription when expressed as GAL4-DBD fusions and the locations of two domains within mSin3A that are able to interact with SMRT. Numbers above the schematic refer to the relevant amino acid positions. (B) Domains of Sin3A able to repress transcription in a transient-transfection assay. Different domains of mSin3A, as depicted schematically on the left, were fused to a GAL4-DBD-coding frame and expressed in CV-1 mammalian cells. The cells were simultaneously transfected with a reporter plasmid containing an SV40 late promoter bearing five binding sites (GAL4 17-mers) for the GAL4-DBD and driving the expression of a luciferase reporter gene. The cells were harvested 48 h later, and the luciferase activity was determined relative to that of a pCH110 β-galactosidase reporter, lacking GAL4 binding sites, employed as an internal control. Fold repression (right) was calculated as the reduction in luciferase expression mediated by a GAL4-DBD fusion construct relative to that mediated by an empty GAL4-DBD (DBD). The results represent the averages and standard deviations from at least two duplicate experiments. (C) Domains of mSin3A able to interact with SMRT in a mammalian two-hybrid interaction. GAL4-DBD fusions representing different domains of mSin3A, as depicted schematically on the left of the panel, were introduced into CV-1 cells together with the GAL4 (17-mer) luciferase reporter and a series of GAL4-AD constructs. The GAL4-AD constructs included an empty GAL-AD construct, a GAL4-AD–SMRT (codons 96 to 566) construct, a GAL4-AD–SMRT (codons 751 to 1495) construct, a GAL4-AD–SMRT (codons 214 to 474) construct, and a GAL4-AD–SMRT (codons 214 to 336) construct. After 48 h, the cells were harvested and the luciferase activity was determined relative to that of pCH110 employed as an internal control (Relative Luc). The results represent the averages and standard deviations from at least two duplicate experiments.