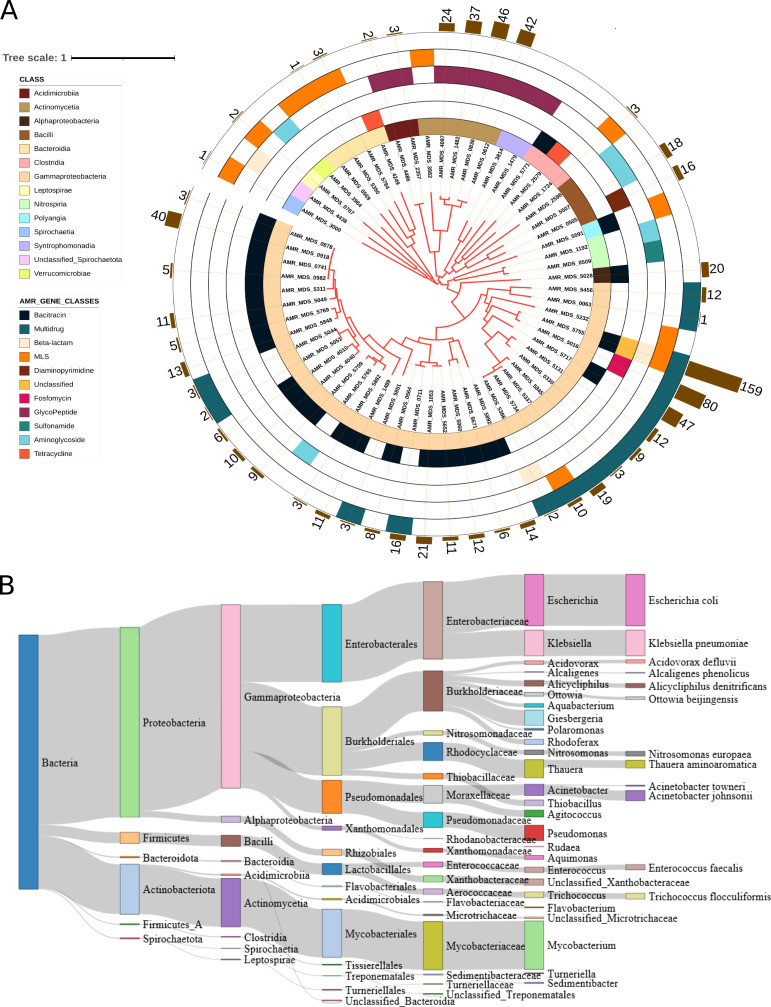
Fig 7.
Phylogenetic tree of the 45 bacterial metagenome-assembled genomes (MAGs) carrying resistomes and host antibiotic-resistant pathogens. (A) Maximum likelihood phylogenetic tree of MAGs constructed using 120 bacterial marker genes in GTDB-Tk. Leaf labels metagenome-assembled genomes sequence. Rings, from the inner to the outside circles, represent Ring 1, which displays the taxonomy of the MAGs at class level, and Rings 2–5, antimicrobial resistance gene classes that are widespread and sparse in bacterial genomes. Bar charts indicate the number of virulence factor genes found in the MAGs with antibiotic resistance genes. The size of the bars shows the number of virulence factor genes detected in MAGs. (B) Range of putative antibiotic resistance pathogens across activated sludge and wastewater samples. The Sankey displays the taxonomy of these putative pathogenic bacteria carrying virulence factor genes. Taxonomy was defined using GTDB-Tk. The size of the bars indicates the relative frequency of potential pathogens.