Fig. 1.
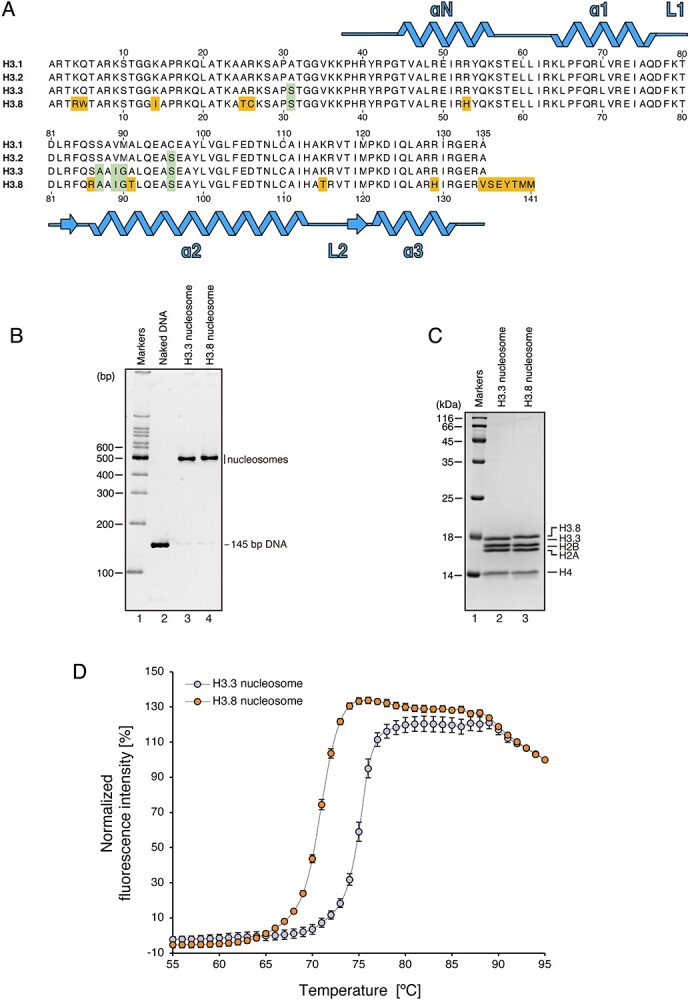
Structural instability of the nucleosome formed with H3.8. (A) Amino acid sequence alignment of human histones H3.1, H3.2, H3.3 and H3.8. Conserved residues between H3.3 and H3.8, but not H3.1, are highlighted by green boxes, and H3.8-specific residues are highlighted by orange boxes. (B) Non-denaturing PAGE analysis of purified nucleosomes containing H3.3 or H3.8 with EtBr staining. (C) SDS-PAGE analysis of the purified nucleosomes containing H3.3 or H3.8 with CBB staining. (D) Thermal denaturation curve of the H3.8 nucleosome. Histone proteins thermally dissociated from the nucleosomes are detected by SYPRO Orange fluorescent dye, which hydrophobically binds to the surface of the denatured histones. The error bars indicate standard deviations (n = 3). Orange and blue circles represent experiments with the H3.8 nucleosome and H3.3 nucleosome, respectively.
