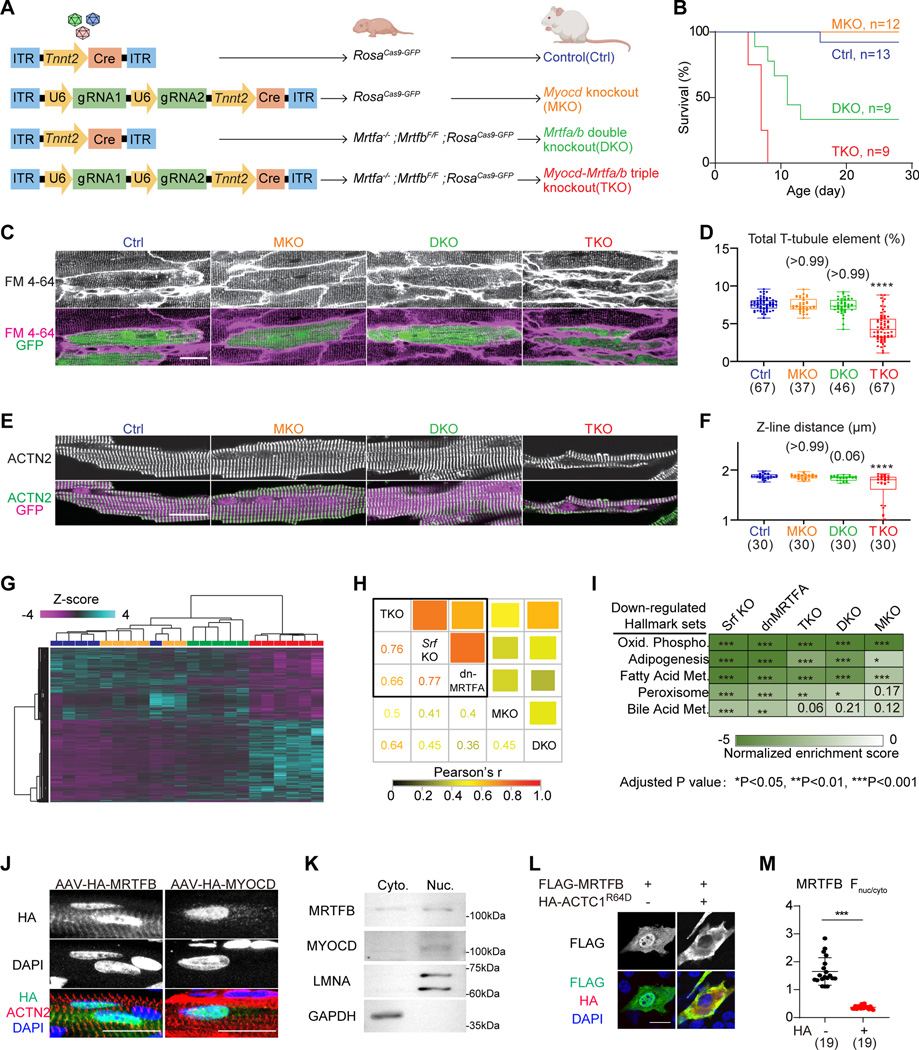
Figure: A shared role of the myocardin-family transcriptional coactivators in cardiomyocyte maturation.
(A) A schematic diagram showing the production of mice with different mutations of the myocardin-family genes. (B) Survival curves of mice that were injected with 3×109 AAV particles at P1. (C) In situ confocal imaging of hearts that were perfused with the cell membrane dye FM4–64. (D) T-tubule organization was quantified by AutoTT software. (E) Fluorescence confocal imaging of isolated cardiomyocytes immunolabelled with ACTN2 antibody to detect sarcomere Z-lines. (F) Distance between adjacent Z-lines was quantified. (G) Hierarchical clustering analysis of differentially expressed genes identified by RNA-Seq. (H) Pair-wise correlation among MKO, DKO, TKO, the published Srf KO and dnMRTFA data for the union of all differentially expressed genes. Both color and square size represents Pearson’s r. (I) Heatmap of top down-regulated hallmark gene sets.
(J) Confocal images of P14 immunostained heart sections that were treated with AAV-MRTFB or AAV-MYOCD at P1. (K) Western blot analysis of endogenous MRTFB and MYOCD in cytoplasmic (Cyto.) versus nuclear (Nuc.) fractions of a wild-type heart. (L) Immunofluorescent staining of NIH3T3 cells that were treated with the indicated plasmids. (M) Quantification of nuclear versus cytoplasmic fluorescence intensity. Scale bars indicate 20 μm in all relevant panels. Box-and-whisker plots: horizontal lines indicate 25th, 50th and 75th quantiles; whiskers extend to maximum or minimum values. Kruskal-Wallis test with Dunn’s multiple comparison correction: *Padj (adjusted P-value)<0.05, ***Padj (adjusted P value)<0.001, ****Padj (adjusted P value)<0.0001, non-significant Padj value in parenthesis above the whiskers. The number of measured cells were indicated in parenthesis below the plots.