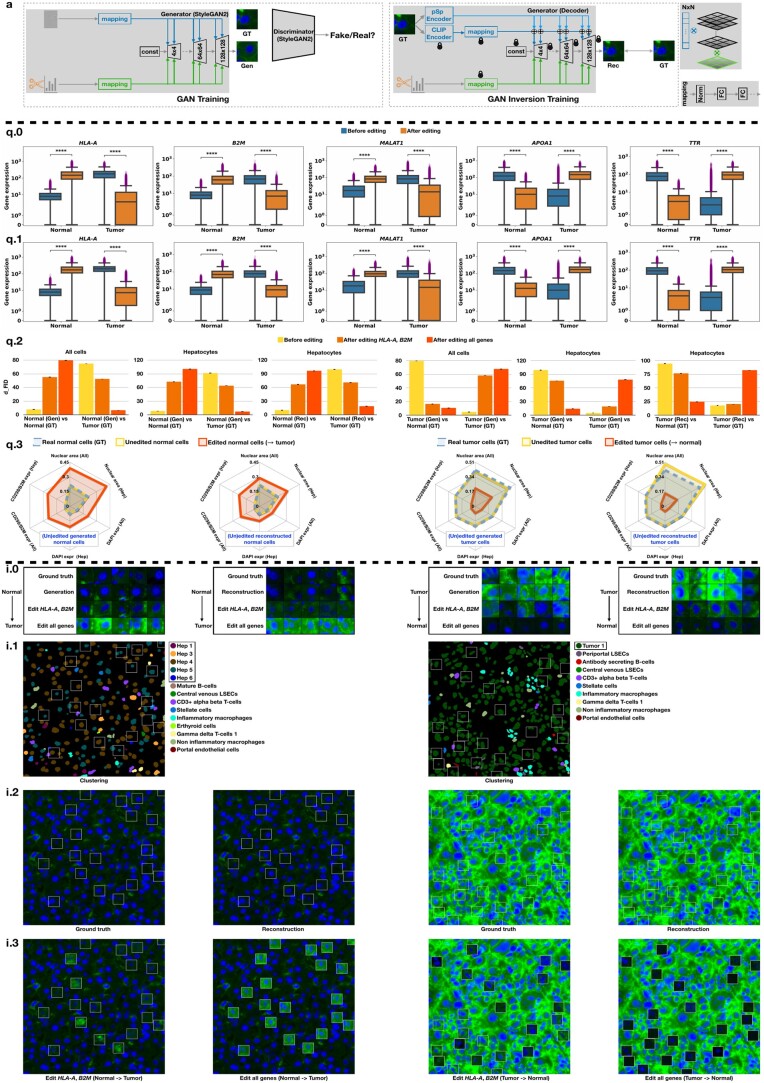
Figure 1.
(a) Model illustrations. (q) Numerical quantification of CosMx results. (q.0 and q.1) The comparison of gene expression shifts on all the cells (q.0) and the “Hepatocyte” (Hep) subtypes (q.1) of the normal and tumor slide. Here, **** means P ≤.0001. (q.2) For the generated (Gen, GAN) and reconstructed (Rec, GAN Inversion) cells, the comparison of cellular state transitions w.r.t. all cells and Hep subtypes. We randomly repeat the computation four times and report the mean and standard deviation. (q.3) The editing effect comparison of interpretable cellular features for all the cells and hepatocytes (Hep). (i) Visual interpretation of CosMx results. (i.0) The image gallery of cellular state transitions for Gen and Rec cells. Here, we present the transition that occurred in the DAPI (blue) and CD298/B2M (green) channels. (i.1) The visualization of cell subtypes on a region of interest extracted from the normal and tumor slide. (i.2) The randomly sampled ground truth and reconstructed cellular images within the bounding boxes. (i.3) The morphological transitions of these cellular images guided by edited gene expression levels