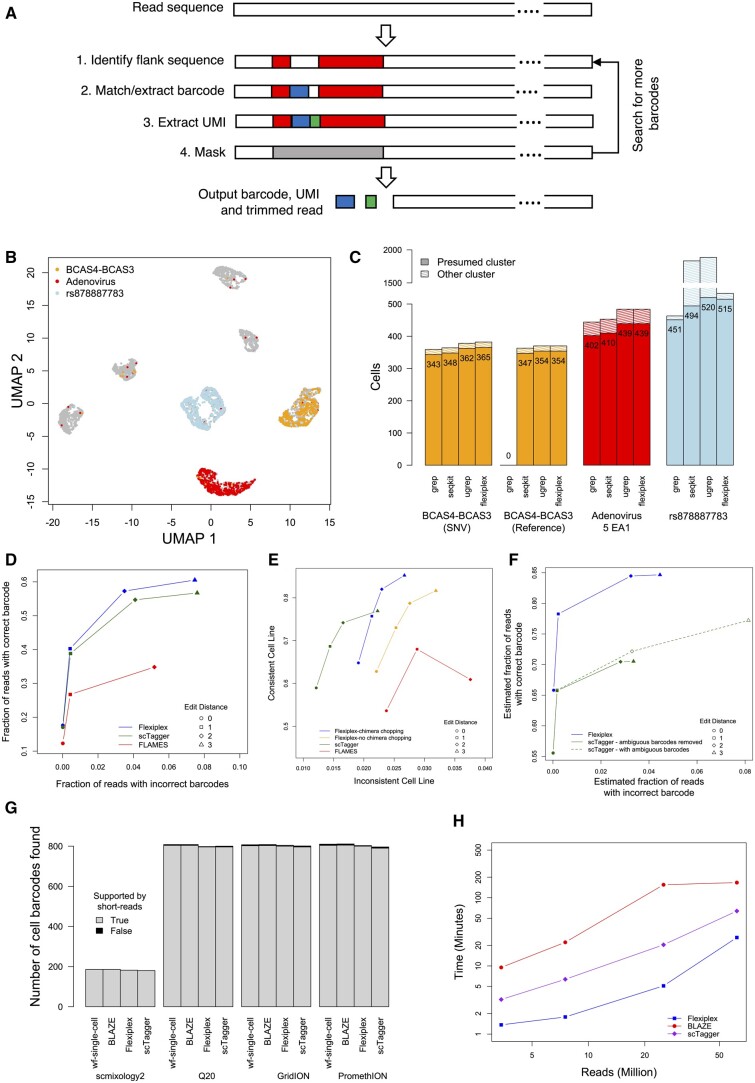
Figure 1.
(A) The demultiplexing approach used by Flexiplex. The right and left flank are first searched for within a read. The barcode and UMI regions are then extracted from the intermediate sequence, with barcode error correction if known barcodes are provided. (B) UMAP of the short-read single-cell dataset of seven pooled cell lines. Cells positive for BCAS4-BCAS3, Adenovirus 5 EA1, and rs878887783 are indicated. (C) The number of cells identified with grep, seqkit grep, ugrep, and Flexiplex that express sequence from BCAS4-BCAS3 (SNP—using an MCF-7-specific variant or Reference—using the reference allele), Adenovirus 5 EA1, and rs878887783 in a short-read single-cell dataset of seven pooled cells lines. Cells, which cluster away from the presumed cluster (hatched), are likely to be false positives, whereas those falling within the presumed cluster are true positives (values on bars). (D) The accuracy of barcode demultiplexing on a simulated set of 5 million single-cell RNA-seq long reads for Flexiplex, scTagger, and FLAMES, varying the maximum allowed edit distance to known barcodes between zero and three. (E) Assessment of cellular barcode demultiplexing on a real dataset of 248 cells sequenced with ONT for Flexiplex (with and without chimeric read splitting), scTagger, and FLAMES, varying the maximum allowed edit distance to known barcodes between zero and three. Correct barcodes will result in a higher level of consistent cell-line annotation. (F) Performance of Flexiplex and scTagger on a large dataset of 61 million reads, where decoy barcodes were used to assess demultiplexing accuracy. As scTagger reports multiple barcodes of equi-distance for each read, we assessed its performance by either removing reads with ambiguous reads, or counting any true barcode as a true positive. (G) The number of barcodes recovered across four datasets when no known barcode list was provided. As scTagger does not adjust the produced barcodes to remove empty droplets like the other methods, we used a script provided with Flexiplex, flexiplex-filter, to automatically refine the barcodes based on the end of the inflection point of the read-barcode frequency distribution. (H) The run-time (log scale, four threads) of stand-alone tools for barcode discovery, Flexiplex, BLAZE, and scTagger, as a function of the number of reads processed from the four datasets used for barcode discovery evaluation. See text and Supplementary Material for further details.