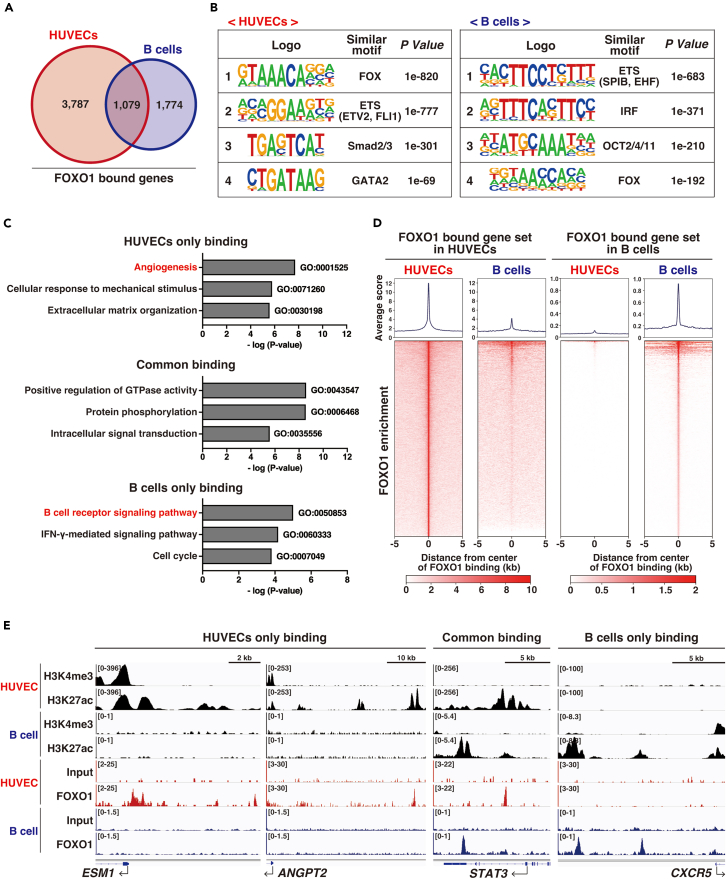
Figure 6.
FOXO1 expressed in ECs preferentially bind to EC-selective expressed genes
(A) Venn diagram depicting the overlap of FOXO1 binding genes between HUVECs and B cells.
(B) MEME co-enrichment analysis of FOXO1 ChIP-seq results from HUVECs (left) and B cells (right). The position-weight matrix indicates enriched sequences. The p value indicates the probability that the de novo enriched sequences obtained from ChIP-seq matched to the known consensus motifs by chance.
(C) HUVEC- or B cell-specific and common enriched gene sets are subjected to Gene Ontology (GO) analysis. In each case, up to three GO terms with the highest significance and the corresponding p values are shown in the graph.
(D) FOXO1 enrichment patterns and the scores in HUVECs (left) and B cells (right) around FOXO1-bound regions (±5 kb) in each cell type are shown. Heatmap density around the same genome loci is shown in the lower panel.
(E) Integrated genome view of the representative HUVEC- or B-cell specific and common FOXO1 binding patterns. H3K4me3 and H3K27ac ChIP-seqs are shown as the proximal active promoter and enhancer mark, respectively. Input indicates the negative control for FOXO1-ChIP-seq from each cell.