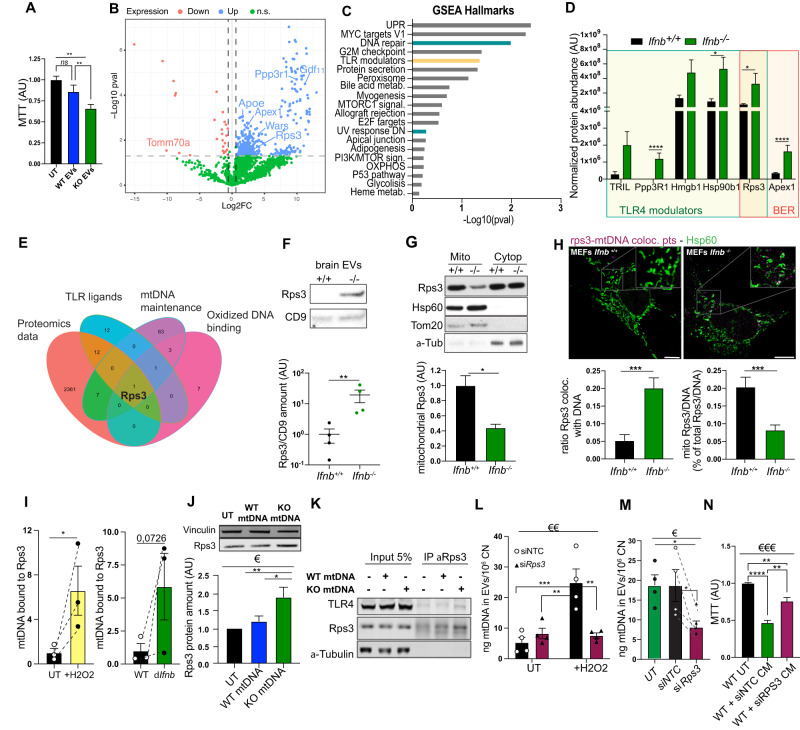
Fig. 6. TLR4 activator Rps3 recognizes damaged mtDNA and is essential for its extrusion in EVs.
A Metabolic activity of CN upon treatment with EVs purified for either wild type or Ifnb−/− CN CM. N = 4. B Volcano plot of proteins differentially expressed in EVs purified from wild-type and Ifnb−/− CN. DEP analysis of N = 3. C Top GSEA pathways dysregulated in Ifnb−/− EVs. D Normalized abundances of some TLR4 modulators and BER proteins found in Ifnb−/− EVs. E Venn Diagram of the common proteins between our dataset, TLR4 ligands, mitochondrial DNA maintenance and oxidized DNA binding pathways. F Immunoblot for Rps3 and EV marker CD9 in EVs purified from wild-type and Ifnb−/− mouse brains. G Cellular fractionation of CNs followed by immunoblot for Rps3. Hsp60 and Tom20 indicate the mitochondrial fraction and α-tubulin the cytoplasmic fraction. H Rps3/DNA colocalization points extracted from immunofluorescence for DNA, Rps3 and Hsp60 (mitochondria) with quantifications in Ifnb+/+ and Ifnb−/− MEFs. N = 3. I Amount of mtDNA CoIP with Rps3 in N2A cells with or without oxidative stress induced by H2O2 or by lack of Ifnb. N = 3. J Immunoblot showing the amount of Rps3 in wild-type CNs treated with either WT or KO mtDNA. N = 3. K Immunoblotting showing CoIP of TLR4 with Rps3 in N2A cells untreated or treated with WTmtDNA or KOmtDNA. L Quantified mtDNA in EVs purified from WT CN ± H2O2 treatment with or without Rps3 knock-down. M Amount of mtDNA quantified in EVs purified from Ifnb−/− CN with or without Rps3 knock-down. N = 4. M Metabolic activity assessed by MTT assay in wild-type cell treated with CM from δIfnb N2A with or without knock-down for Rps3, N = 3. For all graphs, 1 dot means 1 individual animal. €€ means p < 0.01 and €€€ p < 0.001by ordinary one-way ANOVA, Krustal–Wallis ANOVA if distribution did not show Gaussian distribution (Shapiro–Wilk test) or Brown-Forsythe ANOVA if distribution showed SD differences (Bartlett’s test). * means p < 0.05 and ** p < 0.01, by post-hoc unpaired t-test.