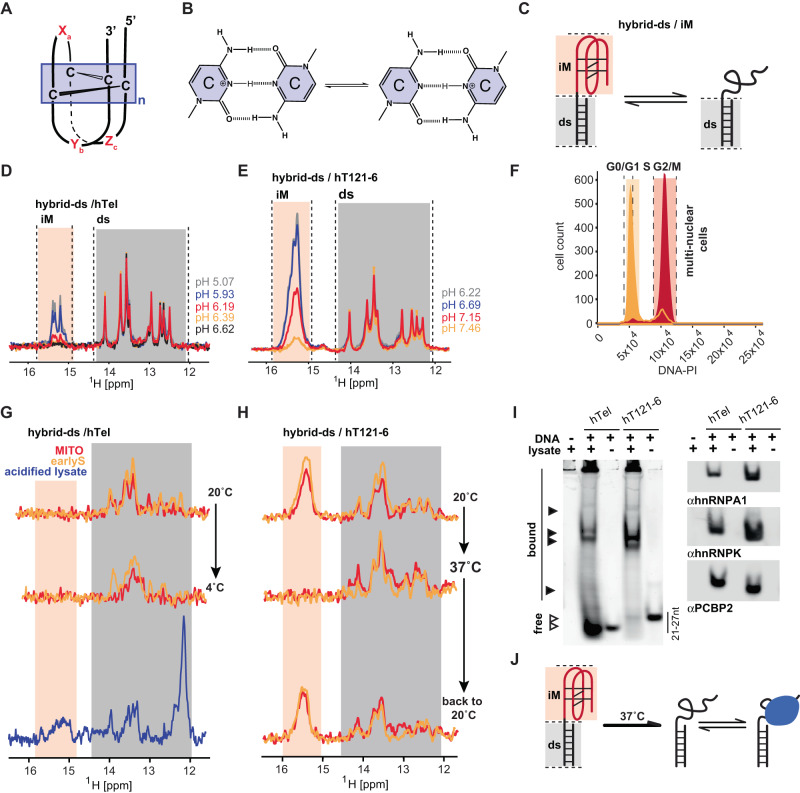
Fig. 1. Hybrid-ds/hTel and hybrid-ds/hT121-6 do not form iM in cells at 37 °C.
A Schematic representations of the DNA i-motif structure with the minimal i-motif segment highlighted in the box. Xn, Yn, and Zn mark i-motif loop regions of variable length. B C:CH+ base-pair, according to ref. 2. C Hybrid-ds/iM reporter of iM-associated structural equilibrium; ds and iM correspond to double-stranded and i-motif segments, respectively. D, E show overlays of the imino regions of 1D 1H NMR spectra of hybrid-ds/hTel and hybrid-ds/hT121-6, respectively, acquired at 20 °C as a function of the pH in-vitro (IC buffer: 25 mM KPOi, 10.5 mM NaCl, 110 mM KCl, 1 mM MgCl2, 130 nM CaCl2). Spectral regions specific for the imino protons involved in C:CH+ (iM) and Watson-Crick (ds) base pairs are indicated in light orange and gray, respectively. F shows propidium iodide (PI) DNA content staining of cells transfected with (FAM-)hybrid-ds/hT121-6 and synchronized in M (red) and early-S (green) cell-cycle phase. G, H show imino regions of 1D 1H in-cell NMR spectra of hybrid-ds/hTel and hybrid/hT121-6, respectively, acquired as a function of the temperature (indicated) in mitotic- (red, MITO) and early-S (green, earlyS) synchronized cells. The displayed in-cell NMR spectra are representative images of two independent experiments. In blue is the NMR spectrum of the hybrid-ds/hTel acquired in the acidified (pH < 6) cell lysate prepared from the respective in-cell NMR sample at 4 °C (folding control). I LEFT: native PAGE of the Cy3-labeled hTel and hT121-6 in the absence and the presence of lysates from HeLa cells visualized via the Cy3-fluorescence. RIGHT: immuno-stained electroblots (PVDF membrane) of the native PAGE with αhnRPNP A1, αhnRNP K, and αPCBP2. The gel and blot are representative images of three independent experiments. Source data are provided as a Source Data file. J Projected representation of structural equilibria involving hybrid-ds/iMs in living cells.