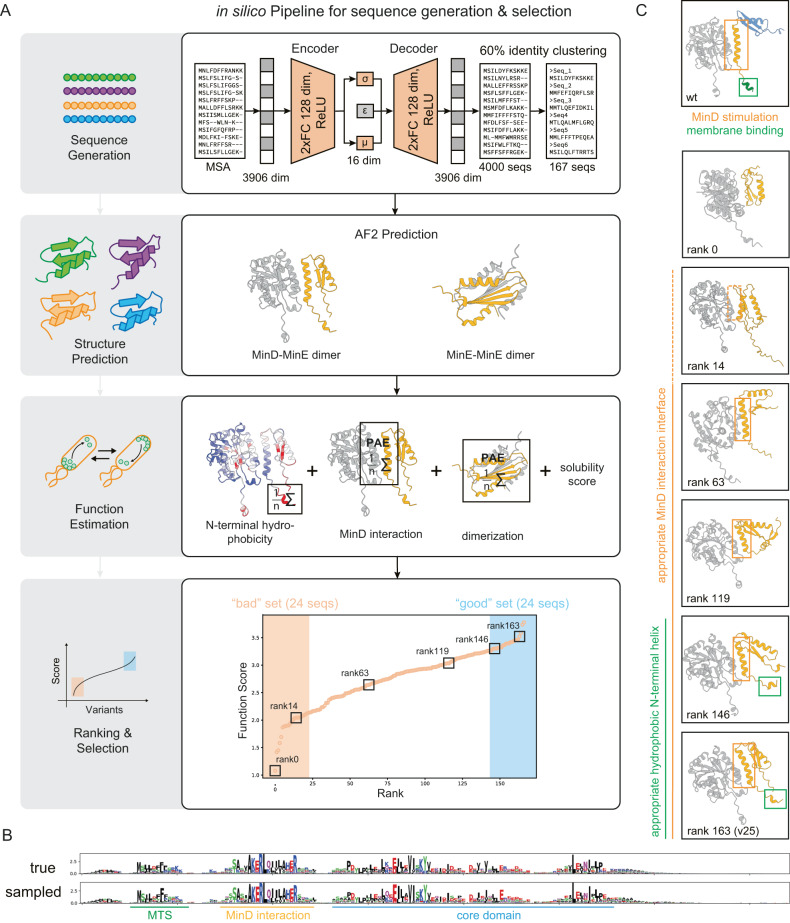
Fig. 2. Generation of MinE homologs and in silico screening for expected function.
a Pipeline overview. Sequences are generated using a Variational Autoencoder and clustered by 60% identity to ensure heterogeneity. The structures of the remaining 167 sequences are predicted using AlphaFold2 for homo- and heterodimers. A function score is calculated based on solubility and the three sub-functions known to allow MinE to oscillate in E. coli: N-terminal membrane binding, interaction with MinD, and dimerization. The results are ranked, and the best and worst 24 candidates are selected for in vitro analysis. b Sequence conservation in naturally occurring and newly generated MinE homologs are highly similar. c Visual validation of ranked structures. With better ranking, structures show better MinD-interaction sites and membrane targeting regions.