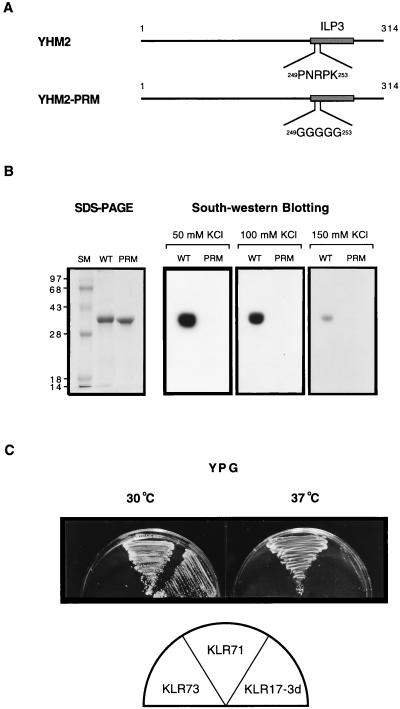
FIG. 6.
The putative DNA-binding domain is essential for suppression of the abf2 mutant. (A) Schematic representation of wild-type (YHM2) and mutant (Yhm2-PRM) Yhm2p. The proline repeat sequence in the putative DNA-binding subdomain was changed to pentaglycine by PCR-based site-directed mutagenesis (see Materials and Methods). (B) The putative DNA-binding subdomain is important for Yhm2p binding to DNA. The wild-type (WT) and mutant (PRM) Yhm2p were analyzed for their DNA-binding activity by Southwestern blotting as described in the legend to Fig. 5B. The same amount of each protein (1 μg) was used for both SDS-PAGE (12% polyacrylamide) and Southwestern analysis. To test the difference of DNA-binding affinity between Yhm2p and Yhm2-PRMp, the concentration of KCl in the binding buffer was varied from 50 to 150 mM. The mutant Yhm2-PRMp did not bind DNA under any conditions. (C) The mutation in the putative DNA-binding subdomain abolishes the suppression of the abf2 mutant. KLR17-3d (abf2::URA3) was transformed with the high-copy-number episomal plasmid harboring the yhm2-PRM gene (pY-D3PRM) and tested for growth on YPG at 30 or 37°C. KLR73 (abf2::URA3, pY-D3PRM) grew very poorly at both temperatures. The control strain, KLR71 (abf2::URA3, pY-D3), grew at both temperatures.