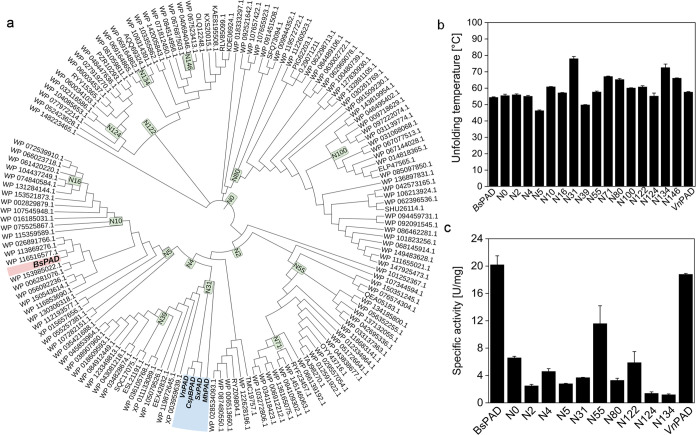
Figure 3.
Ancestral reconstruction of PAD yields a highly thermostable ancestor. (a) Phylogenetic tree composed of 150 sequences of extant phenolic acid decarboxylases. Evolutionary distances were computed with the maximum likelihood method by using the Jones–Taylor–Thornton (JTT) matrix. The phylogenetic tree was visualized using iTOL. Ancestors selected in this study are highlighted in green. (b) The midpoint of the thermal unfolding temperature Tm of each protein was measured by circular dichroism (CD)-spectroscopy at 199 or 224 nm. Conditions: 0.1–0.2 mg/mL of purified protein dissolved in 50 mM KPi, pH 6. The ancestral protein N31 shows higher thermostability in comparison to the extant enzymes from B. subtilis and Vibrio nigripulchritudo. (c) Comparison of the specific activities of ancestors with extant BsPAD toward ferulic acid. Initial reaction rates were measured by following the substrate depletion via HPLC. Reaction conditions: ferulic acid (10 mM, 5% (v/v) dimethyl sulfoxide (DMSO)), 50 mM KPi, pH 6, 2–10 μg of purified enzyme, 30 °C, 600 rpm. Data are the means and range of duplicate measurements. All data were analyzed with Origin 2022b.