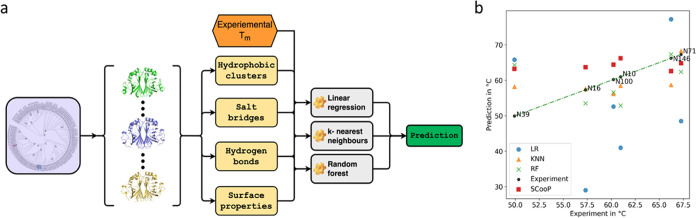
Figure 6.
(a) Overview of the model training and prediction workflow: From the ancestral sequence reconstruction, sequences are selected and expressed, and the experimental Tm is determined. In parallel, AlphaFold 2 models are built from these protein sequences, various structural attributes are calculated, and using these and the experimental Tm as a target, various regression models are fitted to this data. These models can then be used to predict the unfolding temperature of untested ancestors, and these predictions can be used to decide which ancestor to test and which not to test in the lab due to their low predicted Tm. (b) PAD unfolding temperatures: Experimental and predicted values of the three best-performing models and the SCooP server. LR: linear regression, KNN: k-nearest neighbors’, RF: random forest.