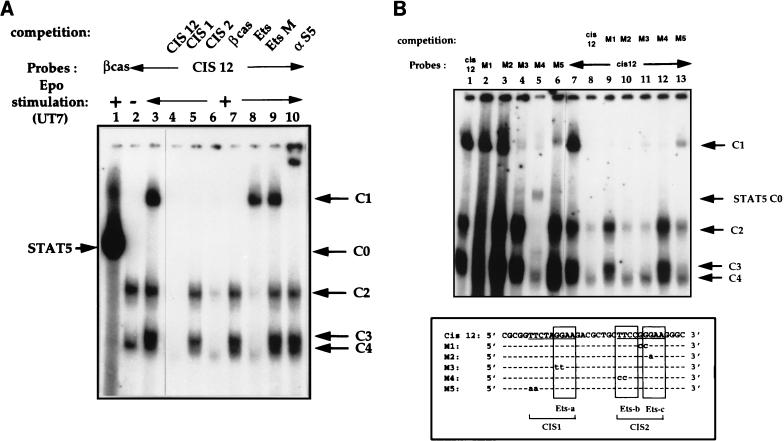
FIG. 6.
Analysis of DNA-protein complexes which bind to the CIS12 probe. Nuclear extracts from Epo-stimulated UT7 cells were incubated with a 32P-labelled CIS12 probe. (A) Identification of specific DNA-protein complexes by competition experiments. Unlabelled competitors (50-fold molar excess) were added to EMSA mixtures as follows: no competitor (lane 3) and double-stranded oligonucleotides CIS12, CIS1, and CIS2; βCAS; an optimized Ets probe from the Drosophila E74 promoter (3); and EtsM, in which the Ets binding sequence GGAA was mutated to CCAA (lanes 4 to 9, respectively). A βCAS probe was used as a control (lane 1). Lane 2 contains nuclear extracts from unstimulated UT7 cells. (B) Mutated CIS12 oligonucleotide bandshift assays. (Top) Nuclear extracts from Epo-stimulated UT7 cells were incubated with the 32P-labelled CIS12 probe (lanes 1 and 7 to 13) and with 32P-labelled mutated CIS12 M1 to M5 probes (lanes 2 to 6, respectively). Competition assays were performed for lanes 8 to 13 with a 50-fold molar excess of probes as indicated in the figure. (Bottom) Alignment of wild-type CIS12 and mutated M1 to M5 sequences. The consensus STAT binding sequence TTCNNNGAA is underlined, and the Ets domains are boxed.