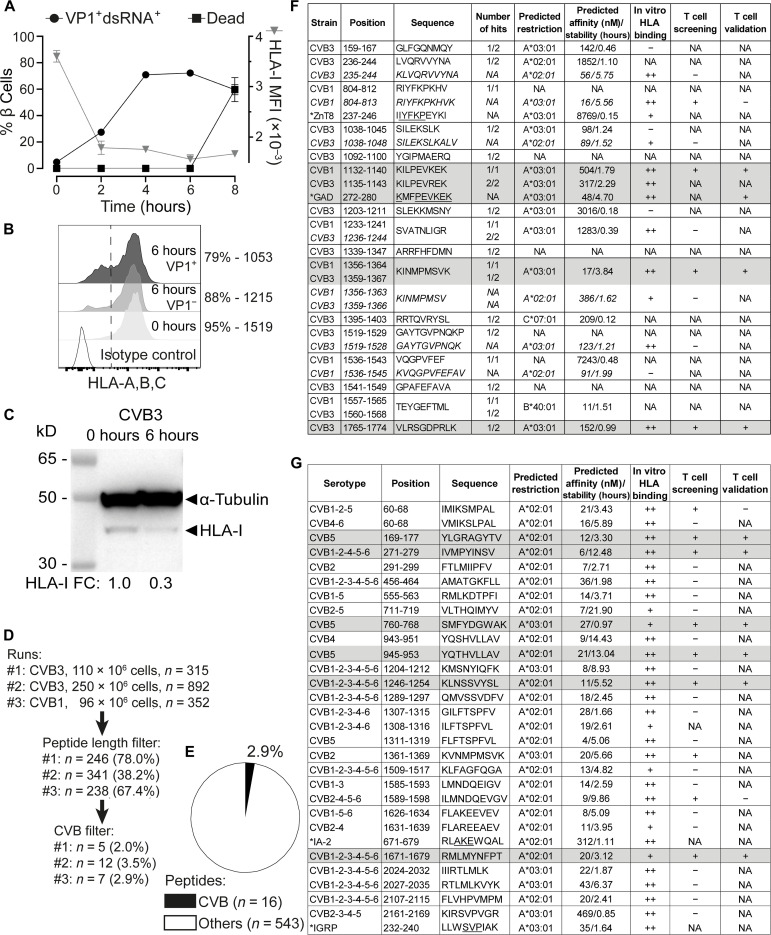
Fig. 1. CVB-infected β cells down-regulate surface HLA-I and present few viral peptides.
(A) Flow cytometry kinetics of viral VP1/dsRNA, cell death (left y axis), and HLA-I median fluorescence intensity (MFI; right y axis) in CVB3-infected ECN90 β cells (300 MOI). (B and C) Flow cytometry HLA-I expression on ECN90 β cells 6 hours postinfection (B; percent HLA-I+ cells and MFI indicated) and Western blot [C; HLA-I fold change (FC) normalized to loading control indicated]. (D) Three immunopeptidomics runs were performed at the 6-hour time point. Sequences were filtered for 8– to 12–amino acid length and for matches with the translated RNA of the CVB strains used; number and percent of peptides retained at each step are shown. (E) Percent CVB peptides out of total 8- to 12-amino acid peptides eluted. (F) CVB peptides retrieved from immunopeptidomics experiments (length variants of identified sequences in italics). The number of hits out of three peptidomics runs are listed for each peptide; see fig. S1 for mapping in the CVB polyprotein. Asterisks indicate CVB1804–812-homologous ZnT8237–246 and CVB11132–1140/CVB31135–1143-homologous GAD272–280 peptides (conserved amino acids underlined). NetMHCStabpan predicted affinity and stability values for the assigned HLA-I restrictions are listed. Subsequent columns list in vitro HLA binding results (see fig. S2) and peptides recognized by CD8+ T cells in the screening and validation runs (Fig. 2 and 3). T cell epitopes eventually validated are shaded in gray. CVB11132–1140 and CVB31135–1143 differ only by 1 amino acid and were counted as one peptide (total n = 16). NA, not applicable (or not assigned for predicted restrictions); +/++, positive; −, negative. (G) CVB nonamer peptides identified in a parallel in silico search for all six CVB strains. Asterisks indicate homologous β cell peptides retrieved by Blast. Three peptides not confirmed as binders in vitro were excluded (see fig. S2). For HLA-eluted peptides, sequence annotations refer to the CVB1/CVB3 strains used. For in silico predicted peptides, annotations indicate sequence identities across serotypes.