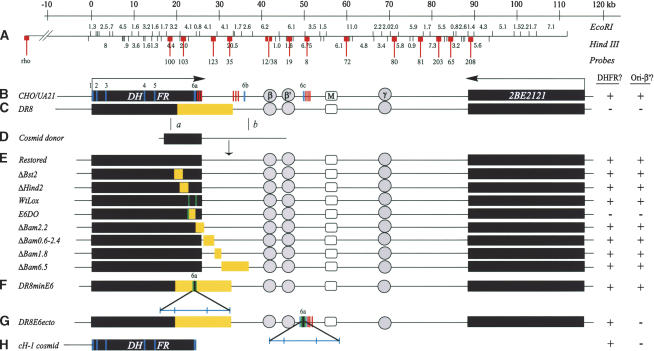
Figure 1.
The DHFR locus in wild-type cells, DR8, and engineered variants. The columns to the right indicate whether an active dhfr enzyme is made in each cell line (DHFR?) and whether replication initiation is detected at a diagnostic locus (ori-β′). (A) Scale and EcoRI and HindIII restriction maps of the wild-type locus in CHO and UA21 cells. Probes used to analyze 2D gels of replication intermediates in various cell lines are numbered and indicated with lollipops (see text). (B) Map of the wild-type locus in diploid CHO and haploid UA21 cells, where black rectangles correspond to the DHFR and 2BE2121 genes and arrows indicate the direction of transcription. DHFR exons are indicated with vertical cross-hatches (note the alternate exons 6b and 6c) and polyA signals with a cluster of three vertical bars. The preferred replication initiation sites in the intergenic spacer (ori-β, ori-β′, and ori-γ) are shown in circles, and a centered matrix attachment site is indicated with a square (M). (C) Map of the DHFR locus in the DR8 variant, where the light-yellow rectangle indicates the region deleted. (D) The KZ381 donor cosmid that is used to restore the locus and/or to introduce targeted deletions by homologous recombination events near a and b. (E). Maps of the various targeted deletions constructed via homologous recombination, where yellow rectangles indicate deletions (see text). (F,G) Variants containing the minimal 168-bp 3′ element inserted either at the deletion junction in DR8 or at the site of the alternative ex6c. Brackets indicate (from left to right) 35 bp lying upstream from the splice site; the 78-bp coding region and stop codon; 55 bp of the 3′ UTR. (H) The cosmid, cH-1, which encodes a functional DHFR gene, and which was transfected into DR8 to test the ability of the dhfr enzyme to restore origin activity. In the table on the right, + and – signs indicate whether the particular cell line expressed a functional dhfr enzyme and whether the ori-β′ region was active.