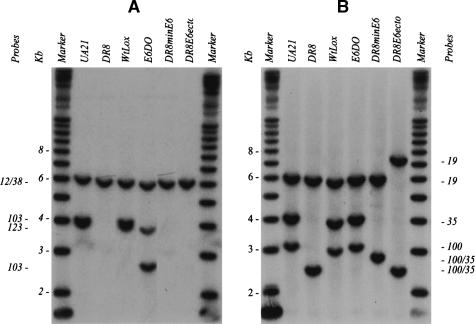
Figure 3.
Southern analysis of diagnostic fragments to confirm the success of targeted deletions and insertions into the DHFR locus. Representative Southern transfers containing EcoRI digests of genomic DNA isolated from the indicated cell lines. Transfers were hybridized with radioactive probe mixtures that illuminate diagnostic fragments for each of the targeted modifications, and each fragment is indicated with the probe that detects it (see Fig. 1A for probe positions and the EcoRI restriction map). (A) In the wild-type hemizygote, UA21, a mixture of probes 12/38, 123, and 103 detects a 6.2-kb EcoRI fragment and a doublet at 4.1 kb. In DR8, only the 6.2-kb EcoRI fragment remains. In WtLox, the pattern is indistinguishable from that of UA21, since the two Lox sites add only ∼60 bp of sequence to the wild-type arrangement. In E6DO, probes 12/38 and 123 detect their cognate wild-type fragments at 6.2 and 4.1 kb, but probe 103 detects a variant of ∼2.6 kb predicted by the deletion of the 1.4 kb encompassing part of intron 5 and exon six. DR8minE6 lacks the wild-type 4.1-kb EcoRI fragments recognized by probes 103 and 123. (B) Additional diagnostic EcoRI fragments were hybridized with probes 19, 35, and 100, and yielded the correct spectrum of fragments in each case (see map in Fig. 1A).