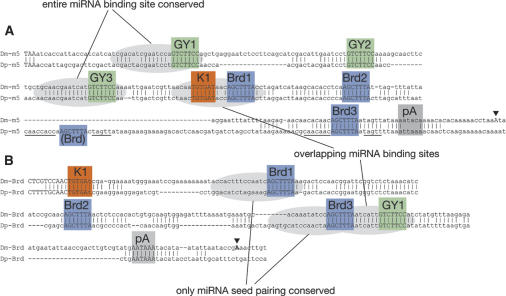
Figure 2.
Alignment of the 3′-UTRs of a bHLH repressor gene [E(spl)m5 (m5)] (A) and a Bearded family gene (Bearded, Brd) (B) from D. melanogaster (Dm) and D. pseudoobscura (Dp). Conserved regions of three or more nucleotides are marked. Polyadenylation sites from cDNAs are labeled with triangles, and polyadenylation signals are labeled pA; note that E(spl)m5 appears to use a noncanonical signal. GY-boxes, Brd-boxes, and K-boxes are all well-conserved between these species but display two classes of divergence: those that preserve the entire miRNA-binding site (box + upstream sequence) and those that preserve only miRNA seed-pairing (i.e., only the box); certain miRNA-binding sites are also overlapping. Note that the sequence surrounding a nonconserved Brd-box in Dp E(spl)m5 is highly related to the sequence surrounding the conserved third Brd-box of E(spl)m5 (cf. nucleotide identities between underlined sequences).