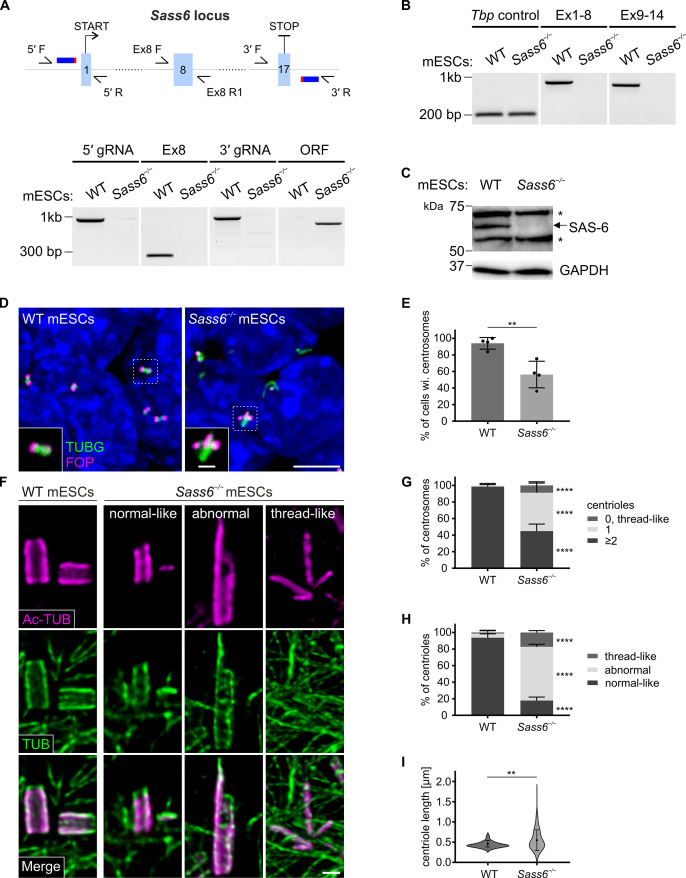
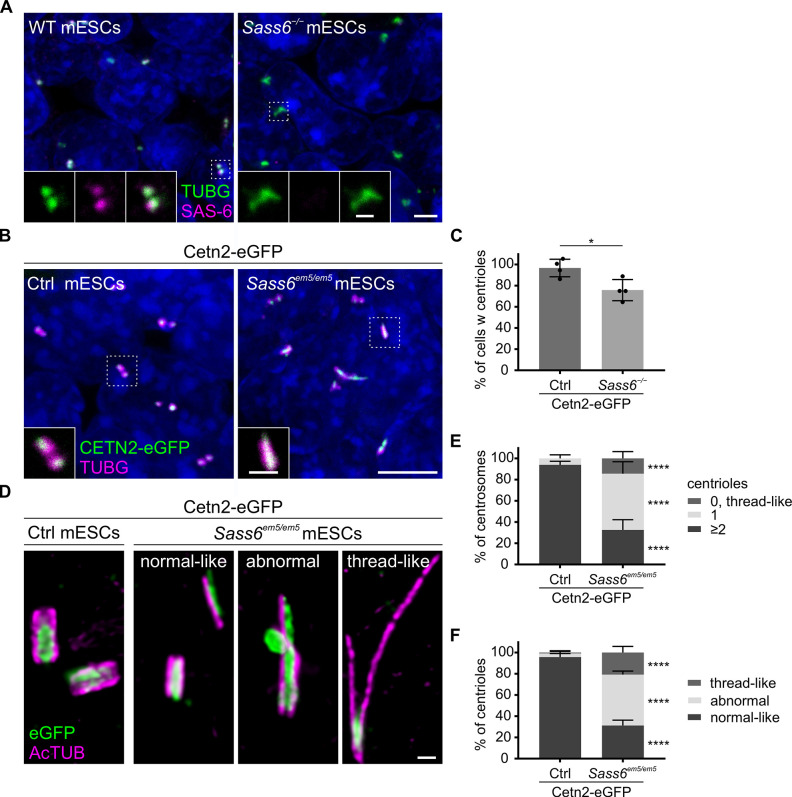
Figure 3. SAS 6 is required for centriole integrity, but not formation, in mouse embryonic stem cells (mESCs).
(A) (Top) Schematic showing the CRISPR/Cas9 strategy using two gRNAs to delete the entire Sass6 open reading frame (ORF) in mESCs. Exons (Ex) are represented by light blue boxes, gRNAs by dark blue thick horizontal lines, and PAM sites in red. Half arrows indicate the primers used for PCR analyses (below). (Bottom) Confirmation of the Sass6 deletion in Sass6−/− mESCs by genomic PCR. The picture shows the PCR products using the following primers indicated in the schematic above: 5′ gRNA (5′ F and 5′ R, band = 977 bp), Ex8 (Ex8 F and Ex8 R1, band = 281 bp), 3′ gRNA (3′ F and 3′ R, band = 992 bp), Sass6 ORF (5′ F and 3′ R, 825 bp in Sass6−/−, 34,349 bp in wild-type (WT), product too long to be amplified). (B) RT-PCR analyses of Sass6 transcripts in WT and Sass6−/− mESCs. The picture shows the PCR products from RT-PCR using the following primers: from Ex1 to Ex8 (Ex1 F and Ex8 R2, band = 734 bp), from Ex9 to Ex14 (Ex9 F and Ex14 R, band = 617 bp), Tbp Ctrl (Tbp F and Tbp R, band = 156 bp). (C) Western blot analysis using a SAS-6-specific antibody on WT and Sass6−/− mESCs extracts. Asterisks mark non-specific bands. GAPDH is used as a loading control. (D) Immunostaining for TUBG and FOP in WT and Sass6−/− mESCs. Insets are magnifications of the center of the dashed squares. Scale bars = 5 µm and 1 µm (insets). (E) Quantification of the percentage of cells with centrosomes (TUBG and FOP) in (D) from four independent experiments. Error bars represent mean ± SD WT: 94 ± 6% (n=2450 cells); Sass6−/−: 56 ± 14% (n=2766). **p<0.01 (two-tailed Student’s t-test). (F) Centrioles were visualized using U-ExM and immunostaining for α- and β-tubulin (TUB) and Ac-TUB in WT and Sass6−/− mESCs. Scale bar = 200 nm. (G) Quantification of the percentage of centrosomes with ≥2, 1, or 0 centrioles in (F) in WT and Sass6−/− mESCs from five independent experiments. Error bars represent mean ± SD WT (n=156 centrosomes):≥2 centrioles = 99 ± 2%; 1 centriole = 1 ± 2%; Sass6−/− (n=254):≥2 centrioles = 45 ± 8%, 1 centriole = 46 ± 10%, 0 centrioles = 9 ± 4%. ****p<0.0001 (two-tailed Student’s t-test) (H) Quantifications of the percentage of centrioles within each category in (F) from five independent experiments. Error bars represent mean ± s.d. WT (n=330 centrioles): normal-like centrioles = 94 ± 4%; abnormal centrioles = 5 ± 3%; thread-like structures = 1 ± 2%; Sass6−/− (n=432): normal-like centrioles = 18 ± 4%, abnormal centrioles = 65 ± 3%, thread-like structures = 17 ± 2%. ****p<0.0001, (two-tailed Student’s t-test). (I) Violin plots of centriole length of normal-like centrioles in (F) in WT and Sass6−/− mESCs from five independent experiments. Error bars represent mean ± SD WT: 0.46 ± 0.07 µm (n=72 centrioles); Sass6−/−: 0.55 ± 0.25 µm (n=72). **p<0.01 (two-tailed Student’s t-test).