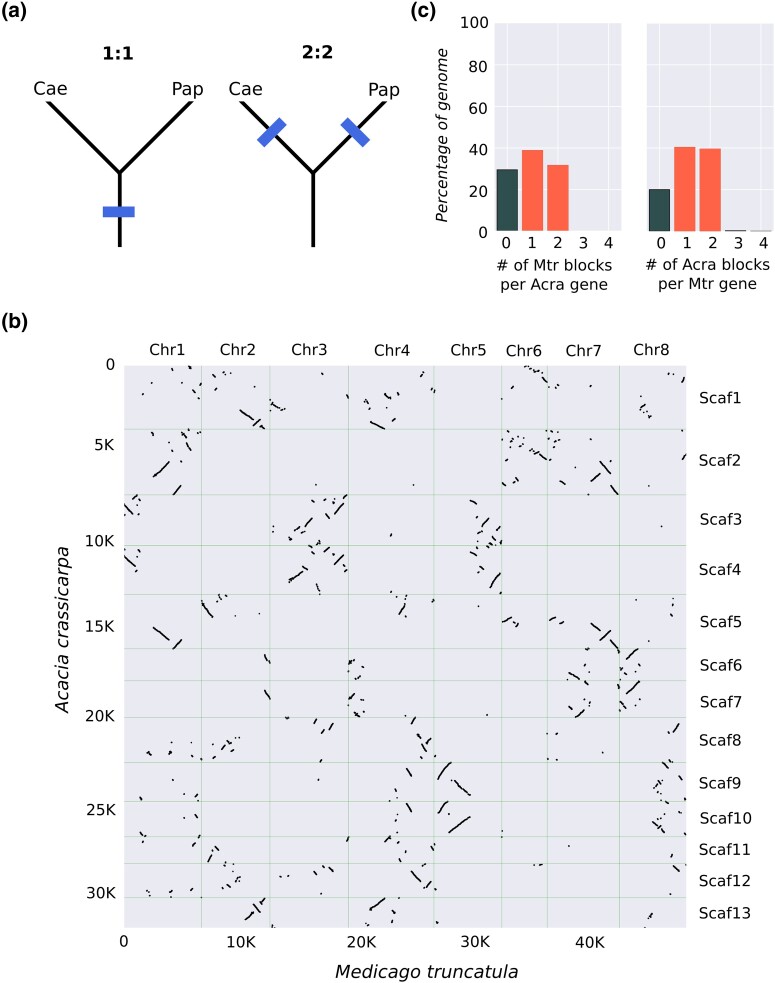
Fig. 2.
A comparison of the A. crassicarpa genome with a M. truncatula. a) Predicted syntenic depth between a Caesalpinoid (Cae) and a Papilionoid (Pap) genome under the 2 leading hypothesis for WGD within legumes. Either the WGD event previously confirmed for Papilionoid species was shared with Caesalpinoid species (left tree) or the event was independent and a second event unique to the Caesalpinoid lineage happened after subfamily divergence (right tree). The former scenario results in a 1:1 ratio between orthologous blocks, while the latter would produce a 2:2 ratio (i.e. 2 orthologous blocks per gene). b) A comparison of the A. crassicarpa genome with M. truncatula. The cumulative number of gene orthologs is plotted on the y- and x-axes as a function of position along the M. truncatula chromosomes (horizontal axes) and A. crassicarpa scaffolds (vertical axes). c) Syntenic depth for orthologous genomic blocks from either the A. crassicarpa or the M. truncatula genome (i.e. the number of orthologous blocks for a gene from the comparison species).