Figure 1.
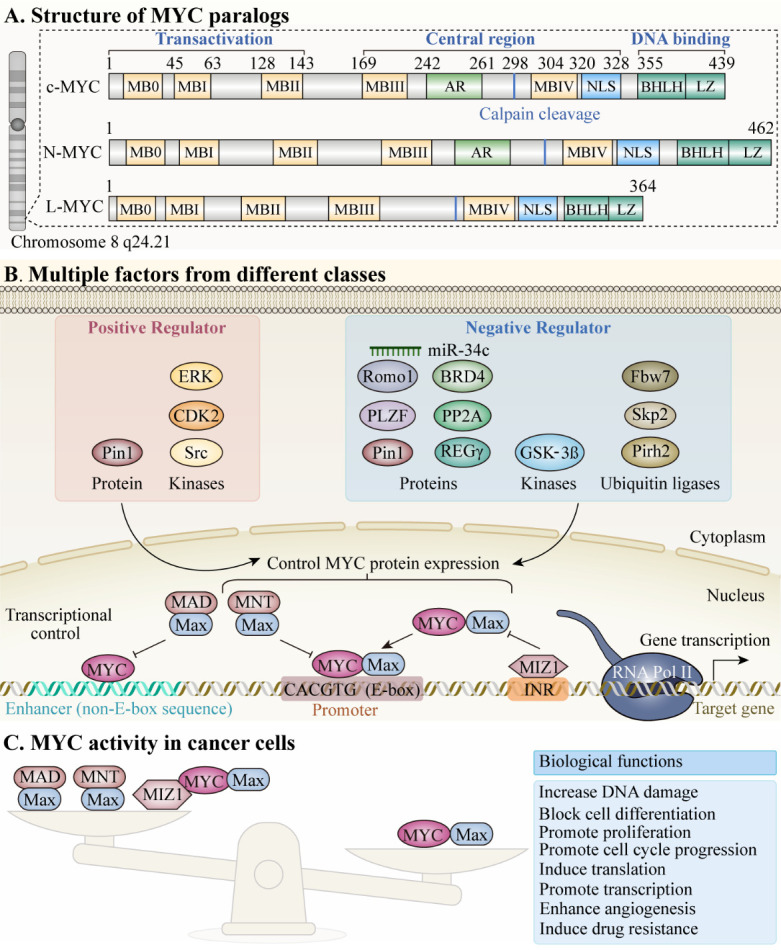
Schematic representations of the functional regions of three MYC paralogs. The N terminus of MYC comprises a transactivation domain locates on residues 1-143 and three highly conserved elements: MYC boxes (MB) 1-3 (MBI: residues 45-63; MBII: 128-143; MBIII: 169-199). FBW7 targets the phosphodegron in MBI. MBII recruits a histone acetyltransferase (HAT) complex that mediates all known MYC functions. MBIII is key to regulating the stability and transcription level of MYC. The BR/HLH/LZ motif (residues 355-439) at the C-terminus is necessary for DNA binding and binding to its canonical partner, MAX. (B) Summary of different classes of positive and negative regulatory factors that influence regulation of MYC expression networks. The transcriptional activities of target genes are driven by the MYC/MAX dimer, which bind to DNA E-boxes or non-E-box sequences. (C) Interactions among MYC, MIZ1, MAX, and MAD (or MNT), different combinations of which exhibit varying effects on transcriptional activity. Disruption of the balance among these complexes can influence significant biological functions and exert cancerous effects in tumorigenesis. BRD4: Bromodomain-containing protein 4; CDK2: Cyclin-dependent kinase 2; ERK: Extracellular signal-regulated kinase; Fbw7: F-box and WD repeat domain containing 7; GSK-3β: Glycogen Synthase Kinase 3 Beta; MAX: MYC Associated Factor X; MIZ1: Myc-interacting zinc finger protein 1; MNT: MAX Network Transcriptional Repressor; Pin1: Peptidylprolyl Cis/Trans Isomerase, NIMA-Interacting 1; Pirh2: p53-induced RING-H2 protein; PLZF: promyelocytic leukemia zinc finger; PP2A: Protein Phosphatase 2A; REGγ: REGgamma proteasome; Romo1: Reactive Oxygen Species (ROS) Modulator 1; Skp2: S-Phase Kinase Associated Protein 2.
