Figure 1.
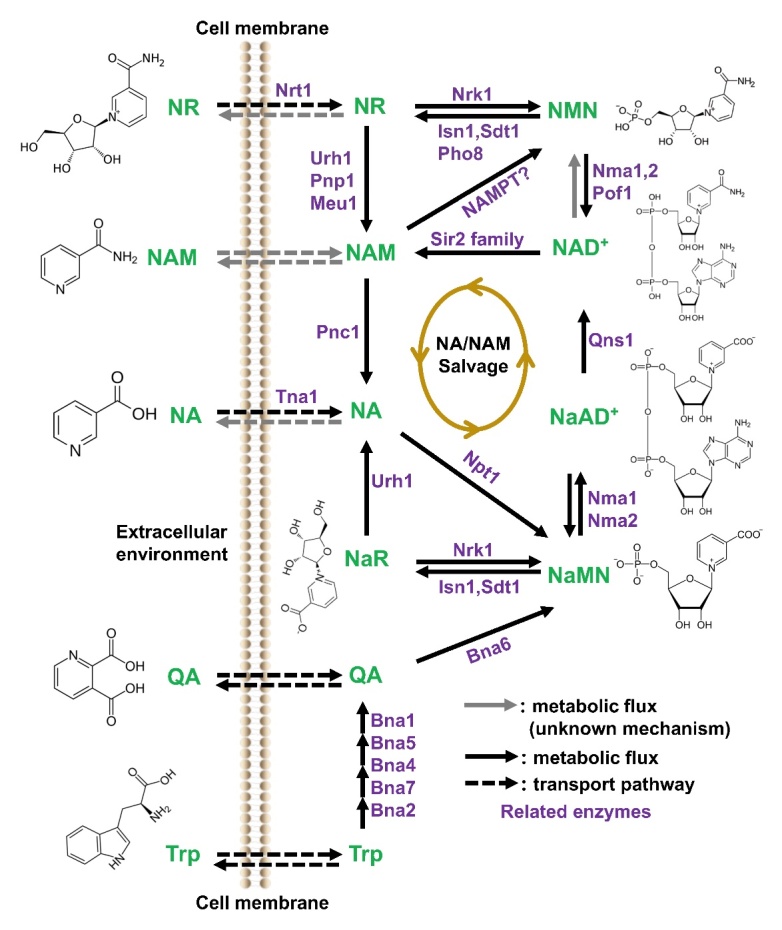
NAD+ and NMN biosynthesis in yeast cells. NAD+ can be synthesized by three pathways: NA/NAM salvage pathways, tryptophan de novo synthesis, and NR-mediated synthesis. Nicotinic acid biosynthesis proteins (Bna 2, 7, 4, 5, 1) are involved in the spontaneous cyclization that leads to the formation of quinolinic acid and NaMN in the tryptophan de novo synthesis pathway. However, this pathway is likely inactive when NAD+ levels are sufficient. In the NA/NAM salvage pathway, NaMN is produced from NA, which ultimately results in the formation of NaAD. The conversion of NaAD to NAD+ is facilitated by Nma1, Nma2, and Qns1. In the NR-mediated NAD+ biosynthesis pathway, NR is first transformed into NMN by the enzyme Nrk1. NMN is then used by the enzymes Nma1, Nma2, and Pof1 to synthesize NAD+. Generally, the Nrk1-dependent and Urh1/Pnp1-mediated routes assimilate NR and utilize exogenous NR made from NMN by the nucleotidase actions of Sdt1and Isn1 in the cytosol. For NMN, rate-limiting enzyme, nicotinamide phosphoribo-syltransferase, converts nicotinamide into NMN. NR kinase (NRK)-mediated phosphorylation is also used to synthesize NMN from NR. NaMN, Nicotinic acid mononucleotide; NA, Nicotinic acid; NR, Nicotinamide riboside; NaAD, Nicotinic acid adenine dinucleotide; NAD, Nicotinamide adenine dinucleotide; NMN, Nicotinamide mononucleotide; Nma, nicotinamide mononucleotide adenylyltransferase; Qns1, Glutamine (Q)-dependent NAD synthetase; NAM, Nicotinamide; Nrt1, Nitrate transporter 1; Urh1, URidine Hydrolase 1; Pnp1, Purine nucleoside phosphorylase; Pnc1, Pyrazinamidase and Nicotinamidase; NAMPT, Nicotinamide phosphoribosyltransferase; Sir2, Silent information regulator 2.
