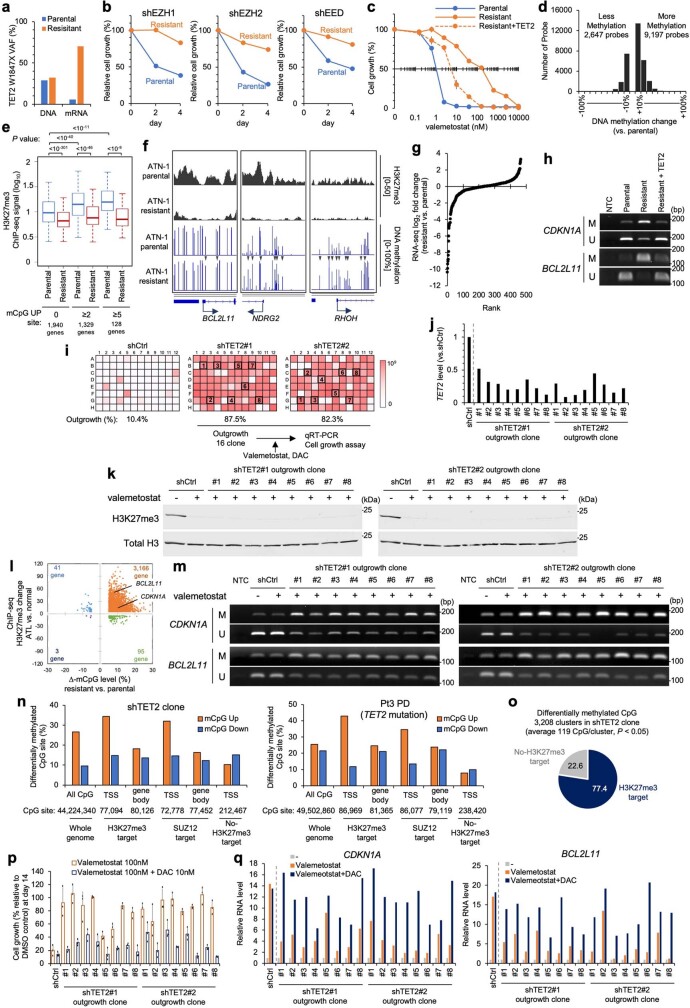
Extended Data Fig. 8. Establishment and characterization of TET2-deficient valemetostat resistant models.
a, ATL cell lines were cultured in growth media supplemented with 10 nM of valemetostat for two months. Inhibitor-resistant outgrowth was observed at 100 nM. Bar graph shows VAF values of TET2W1847X DNA and expressed mRNA in ATN-1 parental and valemetostat-resistant cells. b, shRNA targeting EZH1, EZH2, or EED were introduced by lentivirus vectors in parental and resistant cell line (ATN-1_R). Graphs show cell growth (%) relative to control shRNA. c, Growth inhibition rate (%) by valemetostat (0 ~ 10,000 nM) in ATN-1 parental and valemetostat-resistant cells. n = 3 independent experiments, mean ± SD. For gene rescue experiment, TET2 cDNA were transduced by lentivirus vector. d, Histograms show differentially methylated (ΔmCpG <−10%, or > 10%) probes in resistant ATN-1 versus parental cells. e, f, H3K27me3 occupancy was analyzed by ChIP-seq for the parental and valemetostat-resistant ATN-1 cells. Boxplot shows H3K27me3 log10 signals in relation to resistance-associated mCpG gain (e). The genes for which integrated data were available were evaluated. Statistical significance is provided only for main combinations. Representative tracks for H3K27me3 and methylated CpG tracks are shown in (f). Arrowheads indicate representative CpG sites with methylation gain. g, Log2 fold-changes of RNA-seq expression level (TPM) at mCpG gain genes (mCpG UP sites > 2) in resistant ATN-1 cells compared to parental cells. h, MSP was performed for DNA isolated from parental or resistant ATN-1 cells in the presence or absence of TET2. Amplified DNA was visualized by agarose gel electrophoresis for TSG loci with primer sets specific for methylated state (M) or unmethylated state (U). Data are representative of two independent experiments. NTC: no template control. i, Heatmaps represent recovered outgrowth cell numbers in ATN-1 cells expressing shTET2 (#1, #2) in 96-well plate culture. Collected outgrowth clones (n = 16) are indicated. j, TET2 RNA level in randomly collected outgrowth clones quantified by qRT-PCR. k, H3K27me3 level in valemetostat outgrowth clones. l, Scatter plot shows DNA methylation changes (x-axis) and accumulation of H3K27me3 (y-axis) in the promoter proximal region (TSS ± 1 kbp) of each gene in the outgrowth shTET2 clone #1. Values are averaged per gene and represented only differentially methylated genes (ΔmCpG < −5% or >5%). m, MSP assay for H3K27me3 target genes (CDKN1A and BCL2L11) in valemetostat outgrowth clones. n, Bar graphs show differentially methylated CpG sites in shTET2 outgrowth clone (left) and Pt3 PD clone (right) in single nucleotide resolution analysis using EM-seq data. Percentages were compiled from all CpG sites (filter depth > 5) in the TSS and downstream gene body regions (center ± 1 kbp). Target genes were defined based on H3K27me3, SUZ12, and H3K27ac ChIP-seq data. o, Pie chart shows the percentage of epigenomic domains of CpG islands near the TSS with increased methylation (P < 0.05). p, q, Control cells (shCtrl) and the recovered outgrowth clones were treated with valemetostat (100 nM) and DAC (10 nM). Bar graphs show relative cell growth at 14 days (p, n = 3, independent experiments, mean ± SD) and relative expression levels of the H3K27me3 target genes (CDKN1A and BCL2L11) at 7 days (q). The middle lines within box plots correspond to the medians; lower and upper hinges correspond to the first and third quartiles. The upper whisker extends from the hinge to the largest value no further than 1.5 * IQR. The lower whisker extends from the hinge to the smallest value at most 1.5 * IQR. Statistics and reproducibility are described in Methods. For gel source data, see Supplementary Fig. 1.