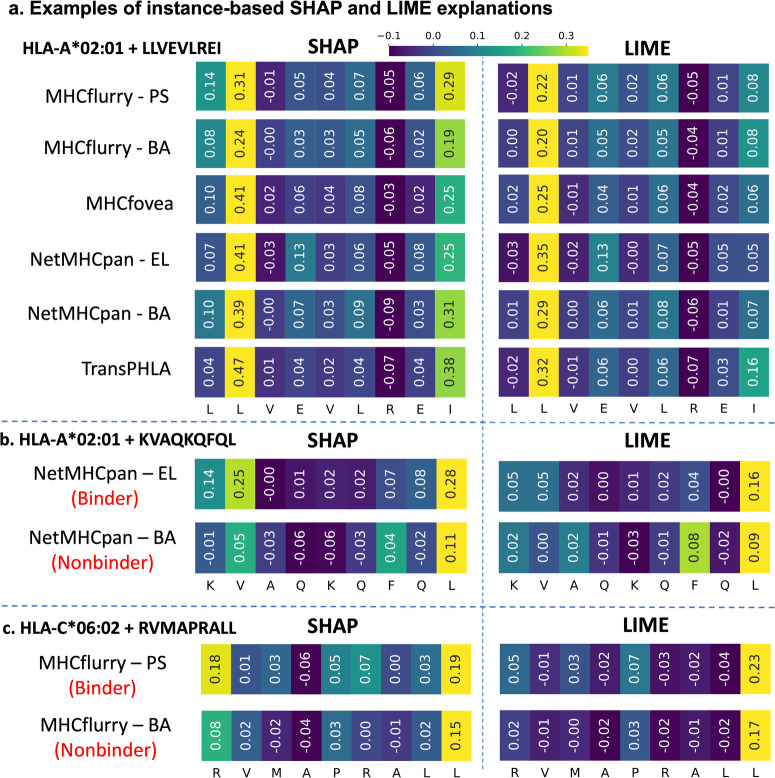
Fig. 3. Instance-based (local) explanations for the Investigated MHC Class I Predictors.
a Examples of SHAP and LIME explanations for all investigated MHC class I predictors for the LLVEVLREI--HLA-A*02:01 pair. To visualize the explanations, the attribution values of positions are used to create heatmaps. For each explanation, a lighter color indicates a positive contribution, while a darker color indicates a smaller or negative contribution to the positive class. SHAP explanations for all the predictors highlight peptide positions P2 and P9 as the most important for binding, while LIME explanations highlight only peptide position P2 as the most important. b LIME and SHAP explanations for NetMHCpan-EL and -BA for the peptide KVAQKQFQL binding to HLA-A*02:01. NetMHCpan-EL classifies KVAQKQFQL correctly, but NetMHCpan-BA does not. SHAP captures these differences in performance and produces different explanations for the two predictor modes. c LIME and SHAP explanations for MHCflurry-PS and -BA for the peptide RVMAPRALL binding to HLA-C*06: 02. MHCflurry-PS classifies RVMAPRALL correctly, but MHCflurry-BA does not. Similar to the example in b, SHAP captures these differences in performance and produces different explanations for the two predictor modes. In both NetMHCpan and MHCflurry examples, LIME explanations are unable to indicate positions leading to the difference in prediction outcome.