Figure 4.
Activation cutoff distinguishes between activated and unactivated macrophages
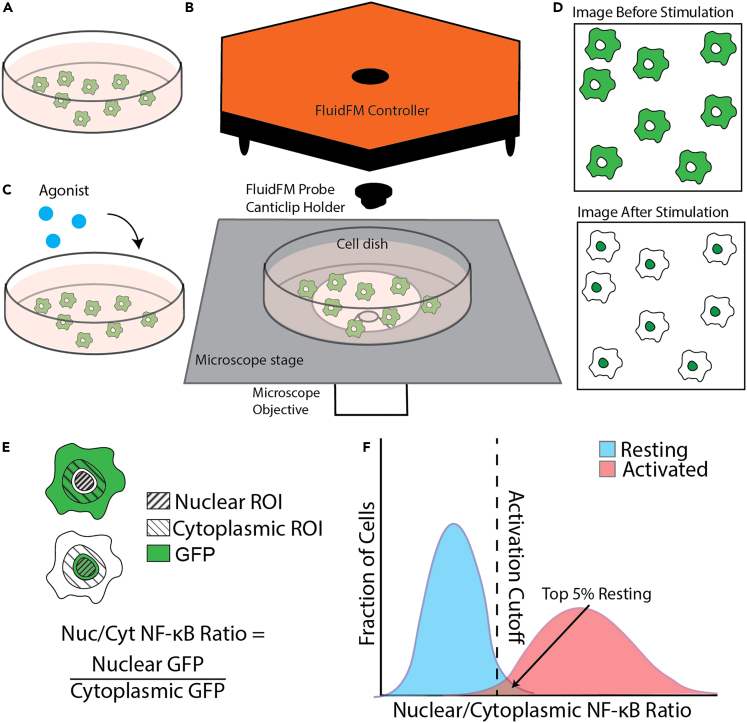
(A) Dish of resting macrophages before stimulation.
(B) Set up for imaging resting macrophages. Include controller and canticlip holder so imaging conditions are the same as for a FluidFM experiment.
(C) Treat macrophages with agonist at a highly activating concentration and image again.
(D) Two sets of images, resting cells before stimulation, and activated cells after stimulation.
(E) Quantify activation of each cell. Nuclear boundaries are drawn from the nuclear stain, then a nuclear ROI is inside that boundary (right diagonal stripes), and the cytoplasmic ROI is a ring just outside the nuclear boundary (left diagonal stripes). This method is most reliable for wide-field images. Measure mean GFP intensity (green) in the two ROIs for each cell. The Nuc/Cyt NF-κB ratio is the ratio of these two measurements. A higher value indicates higher NF-κB activation.
(F) Histogram of the resting and activated macrophage populations. The activation cutoff is set just below the top 5% of the resting population (blue). The power of the cutoff is the percentage of activated cells (pink) above the cutoff. It’s important to make sure there is good contrast between these populations before continuing with quantifying your experiment data, so you can accurately distinguish between activated and unactivated cells. The quantitative cutoff is used to compute the fraction activated metric used in most of the data analysis.