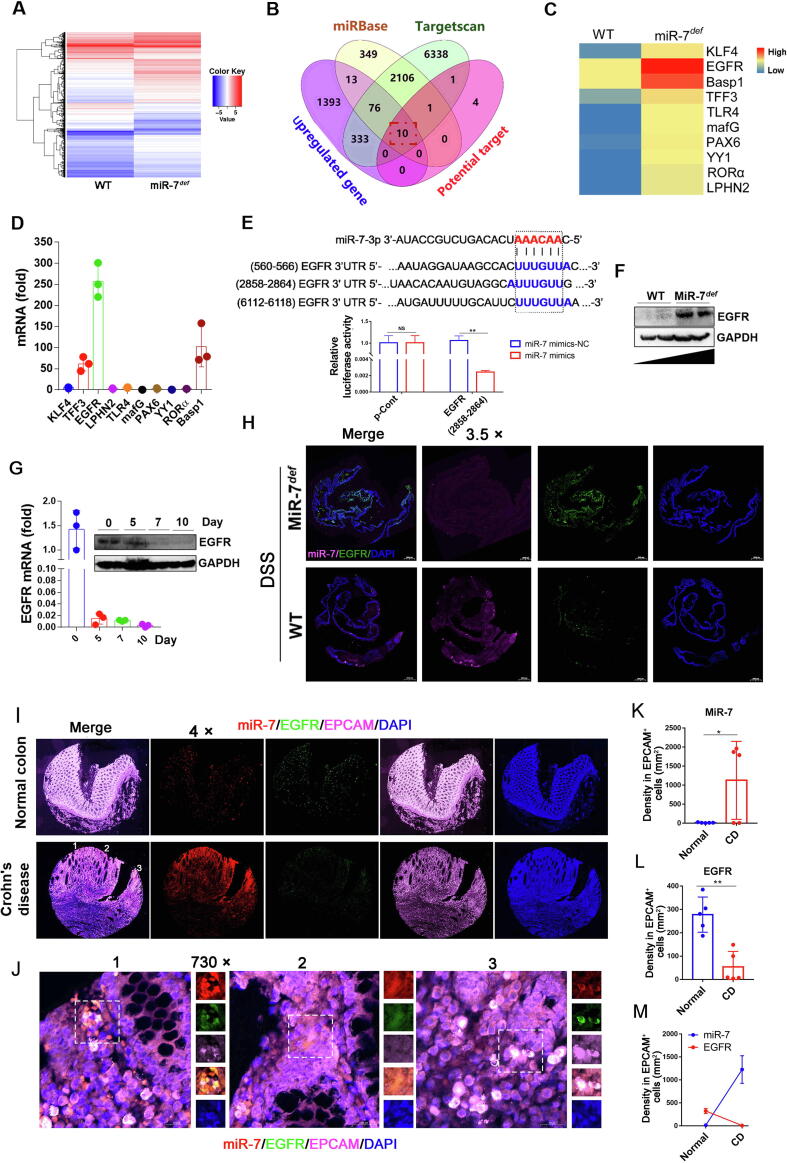
Fig. 5.
EGFR is a target of MiR-7. (A) The gene-expression profiles in the colorectal tissues of miR-7def and WT IBD mice were performed and analyzed using RNA-seq; (B–C) Wenny analysis of miR-7 target genes, whole-genome gene-expression analysis, and related literature; the overlapping 10 genes were screened out as the potential interesting genes. (D) Real-time PCR analysis for KLF4, EGFR, Basp1, TFF1, TLR4, mafG, PAX6, YY1, RORα, and LPHN2 in murine colitis tissue. (E) Putative miR-7 binding sites in the 3′UTR of EGFR from murine species (up); luciferase activity of miR-7 mimics vs mimics NC in WT murine IECs for EGFR (2858–2864 bp site) (down). (F) The protein level of EGFR was detected in the miR-7def and WT murine colitis tissue. (G). The mRNA and protein expression of EGFR in the WT murine colitis tissue were analyzed at indicated time points. (H) The expression of miR-7 and EGFR was detected in colorectal tissues by FISH. The values are the means ± SD (n = 3). (I, K-M) The expression of miR-7 and EGFR were detected in the colon specimens from Normal colon (n = 5) and CD (n = 5) by FISH, then the level and relation of miR-7 and EGFR in EPCAM+ IECs from these specimens were analyzed. Statistical analysis was performed using an unpaired t-test. (J) Representative images of FISH (1, 2, 3) from the CD in Figure I. *P < 0.05, **P < 0.01.