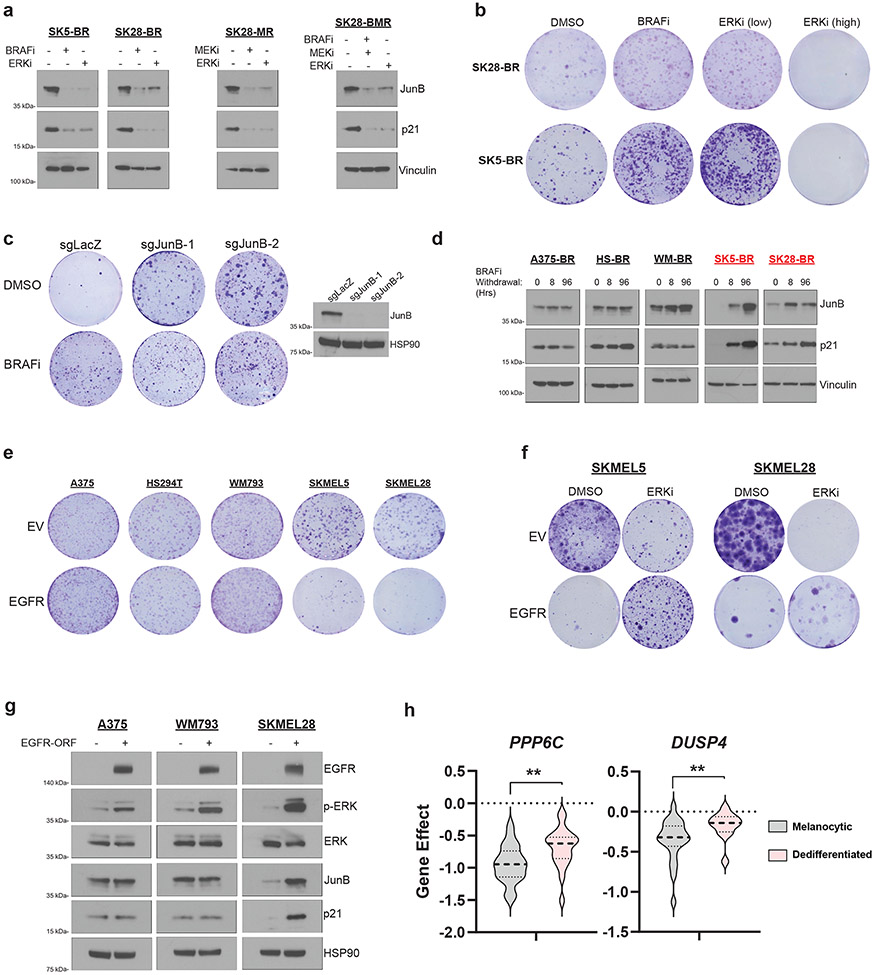
Fig 2: Dedifferentiated melanoma is resistant to ERK-induced cell cycle arrest.
a, Western blot of indicated proteins in SKMEL5-BRAFi Resistant (SK5-BR), SKMEL28- BRAFi Resistant (SK28-BR), -MEKi Resistant (SK28-MR), and -BRAFi + MEKi resistant (SK28-BMR) cell lines treated with the specified doses of DMSO, 3 μM PLX4720 (BRAFi), 3 μM AZD6244 (MEKi), or 50 nM SCH772984 (ERKi) for 96hrs. b, SK28-BR and SK5-BR cells were treated every 72hrs with the indicated drug. Crystal violet staining was performed when each cell line reached confluency. c, (Left) SK5-BR cells with two sgRNAs targeting JunB and one non-targeting control were treated with DMSO or 3 μM PLX4720 every 72hrs. Crystal violet staining was performed on day 14 following seeding. (Right) Western blot validation of JunB knockout. d, Western blot analysis of indicated proteins in BRAFi-resistant A375 (A375-BR), HS294T (HS-BR), WM793 (WM-BR), SK5-BR, and SK28-BR cell lines following BRAFi withdrawal for the specified time. Drug dependent models are highlighted in red. e, Melanoma cell lines with empty vector (EV) or EGFR-ORF construct were seeded in media that contained 2.5 ng/ml of EGF. Crystal violet staining was performed when each cell line's EV condition reached confluency. f, SKMEL5 and SKMEL28 EV and EGFR-ORF transduced cell lines treated with DMSO or 50 nM SCH772984 (ERKi), refreshed every 72hrs. Cells were seeded in media containing 2.5 ng/ml of EGF. g, Western blot analysis of melanoma cell lines transduced with EV or EGFR-ORF following 96 hrs of treatment with 5 ng/ml EGF. h, (Left) Analysis of PPP6C DepMap CRISPR gene score across melanocytic (n = 26) and dedifferentiated (n = 21) MAPK-mutant melanoma. (Right) Analysis of DUSP4 and gene effect across melanocytic (n = 23) and dedifferentiated (n = 12) MAPK-mutant melanoma using the DepMap RNAi (Achilles + DRIVE + Marcotte, DEMETER2) dataset. Two-tailed unpaired t-test, p-values; * <.05 and ** <.01 a-g, The data represent three independent experiments (n =3).