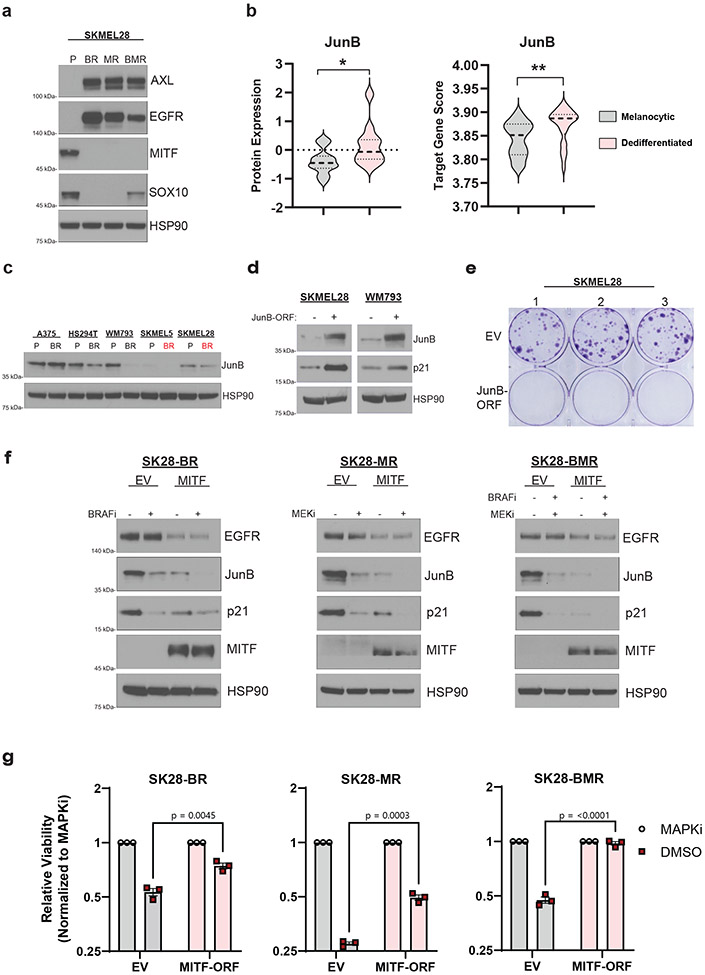
Fig 3: Adaptive loss of MITF is a driver of drug dependence in MAPKi resistant melanoma.
a, Western blot analysis of the indicated proteins in parental (P), BRAFi-resistant (BR), MEKi-resistant (MR), or BRAFi- and MEKi-resistant (BMR) derivatives of SKMEL28 cells. b, (Left) JunB protein level analysis of melanocytic (n = 17) and dedifferentiated (n = 9) MAPK-mutant melanoma cell lines using the DepMap Proteomics dataset. (Right) JunB target gene expression score across melanocytic (n = 36) and dedifferentiated (n = 26) MAPK-mutant melanoma cell lines using the DepMap Expression (Public 22Q4) dataset. Two-tailed unpaired t-test, p-values; ** <.01. c, Western blot analysis of the indicated protein across the specified parental (P) and BRAFi-resistant (BR) cell lines. d, Western blot of SKMEL28 and WM793 parental cells transduced with an empty vector (EV) or JunB expression construction (MOI = 1). e, Clonogenic assay of SKMEL28 cells from experiment d. f, SKMEL28 BRAFi-resistant (SK28-BR), MEKi-resistant (SK28-MR), and BRAFi- and MEKi-resistant (SK28-BMR) cell lines were transduced with an empty (EV) or a full-length MITF (MITF) expression vector and treated with DMSO or the indicated MAPKi for 96 hrs before western blot analysis. g, SK28-BR, SK28-MR, and SK28-BMR were transduced with an empty (EV) or a full-length MITF (MITF-ORF) expression vector. After selection, the cell lines were seeded at 10,000 cells/ml and treated with DMSO or the specific MAPKi therapy to which they evolved resistance. The drug was refreshed every 72hrs. After 10 days, the cells were counted, and the MAPKi counts were normalized to the DMSO counts. Two-tailed unpaired t-test, p-values included in figure. Data are presented as mean ± SEM. a, c-g, These data represent three independent experiments (n =3).