Extended Data Fig. 8 |. rRNA synthesis-dependent localization of DNA-PK in nucleoli of human and mouse cells, and U3 ChIRP-MS of SSU processome.
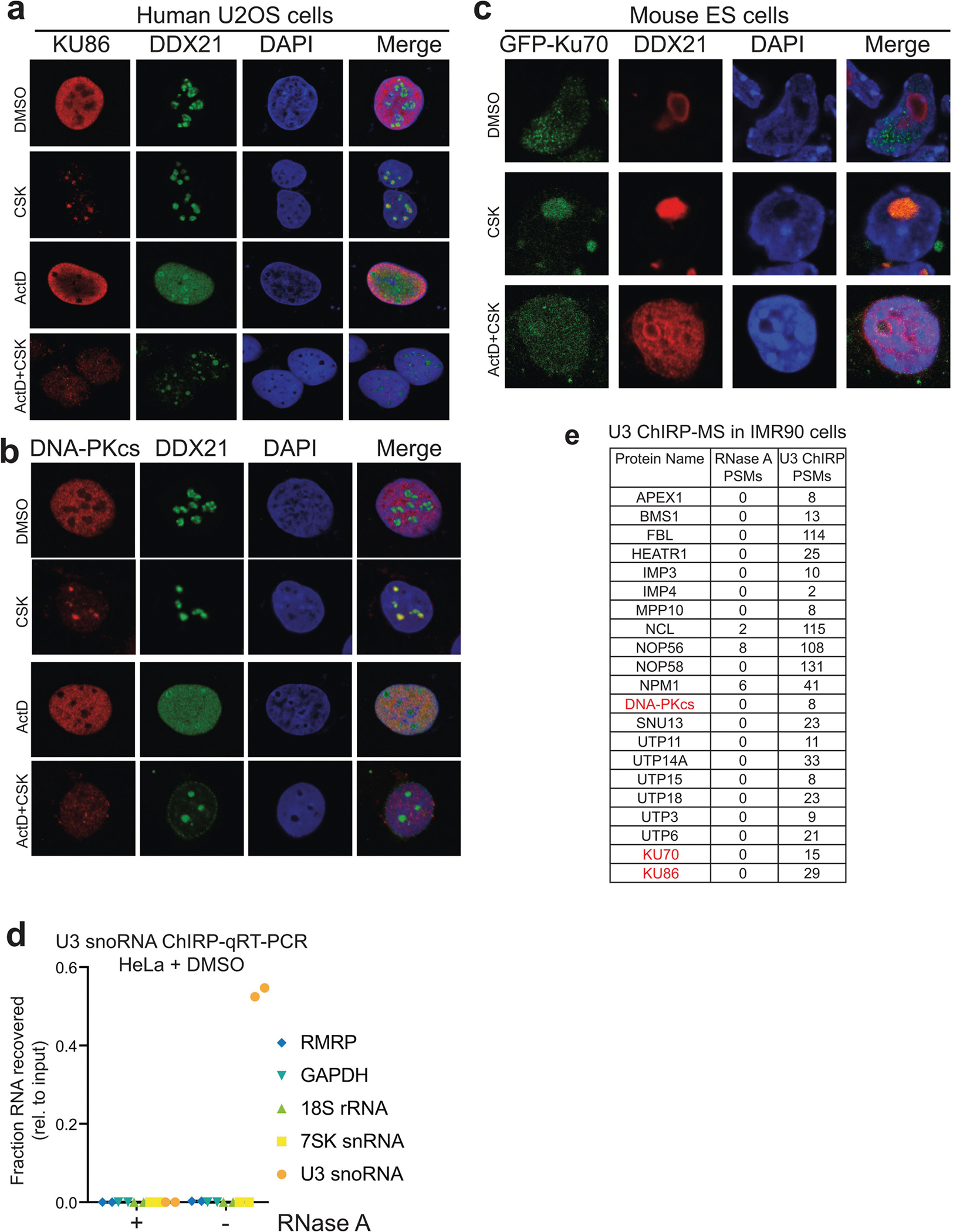
a, b, Immunofluorescence staining of endogenous KU86 (a) and DNA-PKcs (b) in U2OS cells. DDX21 RNA helicase is used as a positive control for nucleoli. The CSK buffer contains Triton X-100 for pre-extraction before fixation (see Methods). When indicated, the cells were treated with 50 nM ActD for 1 h before pre-extraction, fixation and staining. c, Localization of ectopically expressed GFP-tagged KU70 in mouse ES cells. a–c, n = 3 biologically independent experiments. d, U3 ChIRP-qRT–PCR analysis from HeLa cells. Enrichment levels, relative to input samples, of the U3, 7SK, 18S, and RMRP RNAs were assessed from experimental (−RNase A) or control (+RNase A) ChIRP samples. Data are from two independent biological replicates. e, DNA-PK was also recovered from U3 ChIRP-MS in IMR90 cells. Peptide spectral match (PSM) counts for control (RNase A) and experimental (U3) samples are shown. n = 2 biological replicates.
