Fig. 3 |. DNA-PK, but not other cNHEJ factors, co-purifies with the U3 snoRNA and regulates rRNA processing.
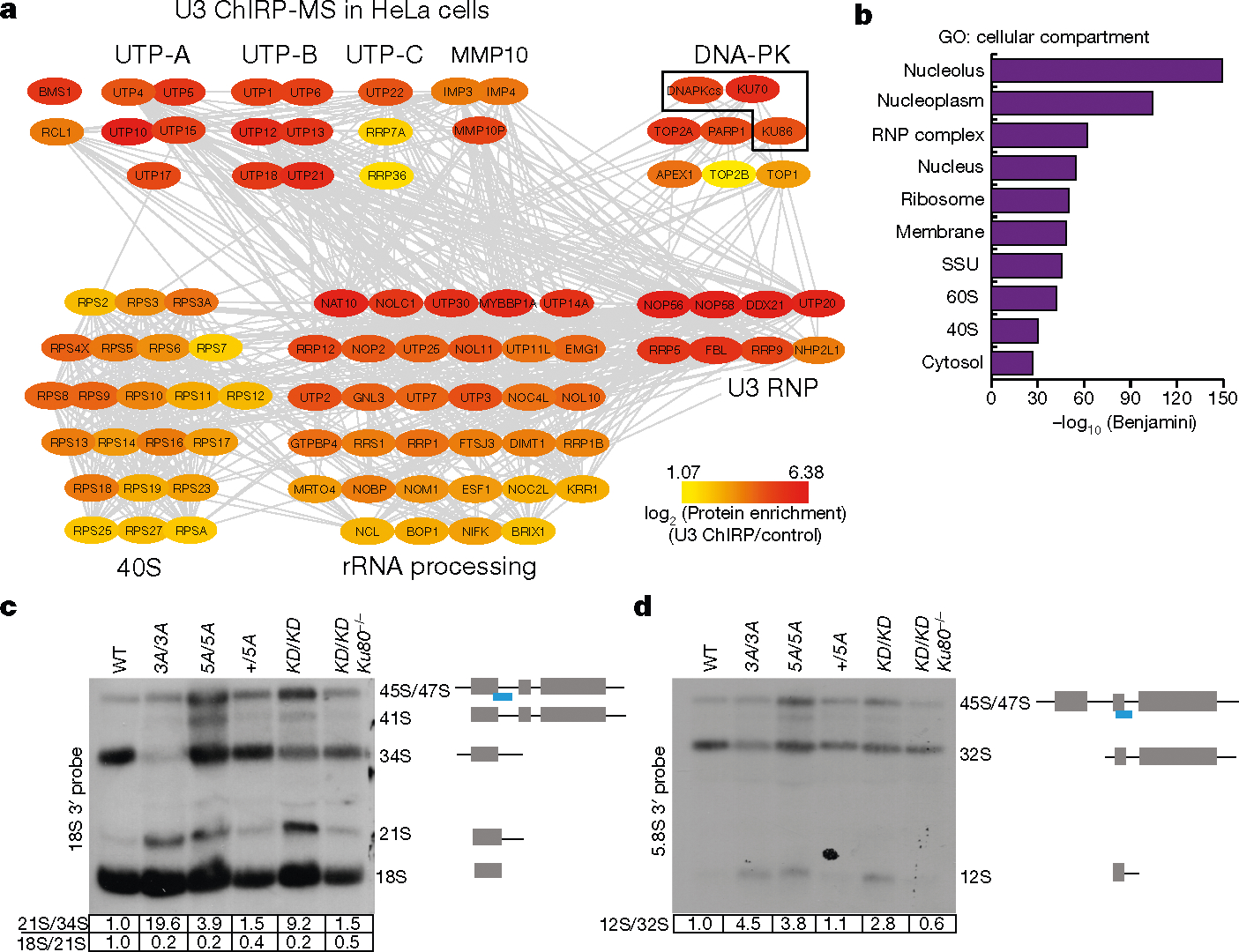
a, A protein–protein interaction (grey lines) network of U3 ChIRP-MS from HeLa cells of nucleolar and ribosomal biogenesis factors. Colours map to the enrichment log2[ChIRP/control] value of each protein. Known complexes are physically grouped and labelled. Complete hit list detailed in Supplementary Table 1. b, Cellular compartment term analysis of the U3 ChIRP-MS from HeLa cells using the DAVID tool. False discovery rate (FDR) was estimated (Benjamini corrected P) by comparing the 483 enriched proteins to the human proteome; the top ten most enriched terms are shown. c, d, Northern blot analyses of 18S and 5.8S rRNA maturation in v-ABL kinase transformed B cells from noted genotypes. The probe covers the 3′ edge of 18S rRNA and 5.8S rRNA, respectively (blue line). The normalized relative intensities of 21S/34S, 18S/21S and 12S/32S are marked below the gels. n = 4 (c), n = 3 (d) biologically independent experiments.
