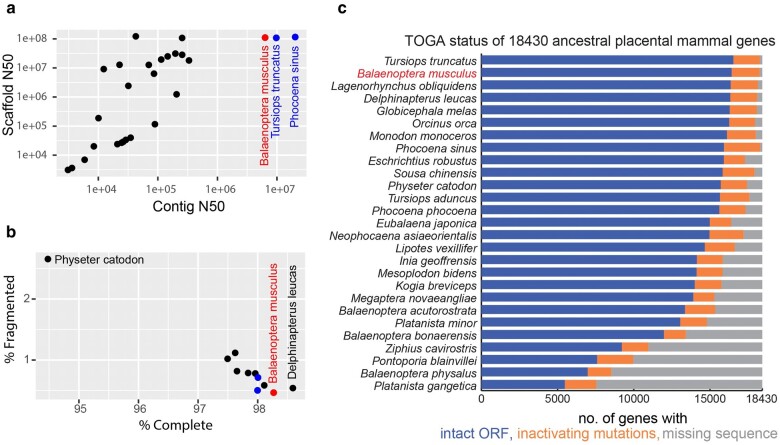
Fig. 1.
Assembly quality metrics. Blue whale (Balaenoptera musculus) data are shown in red; the 2 other VGP assemblies, vaquita (Phocoena sinus) and bottlenose dolphin (Tursiops truncatus), are in blue. a) Assembly contig and scaffold N50 metrics. Contigs are segments of contiguous, i.e. gapless sequence. Scaffolds are sets of contigs that have been ordered and oriented using long-range mapping data such as optical maps and Hi-C with gaps between contigs. N50 is a measure of average length, e.g. 50% of all bases are contained in contigs of length N50 or longer. b) % of complete and fragmented universal single copy BUSCO orthologs found in an annotated genome. Universal single copy orthologs are genes that are present in a single copy in all or most genomes within a phylogenetic group. A high % complete score is an indication that a genome assembly is not missing a large amount of gene-coding sequence (Simão et al. 2015; Manni et al. 2021). C) TOGA status of 18,430 ancestral placental mammal genes. Note: For 2 species, different assemblies were used in panel C compared to panel A: GCA_004363415.1 instead of GCA_002189225.1 for Eschrichtius robustus and GCA_008795845.1 instead of GCA_023338255.1 for Balaenoptera physalus.