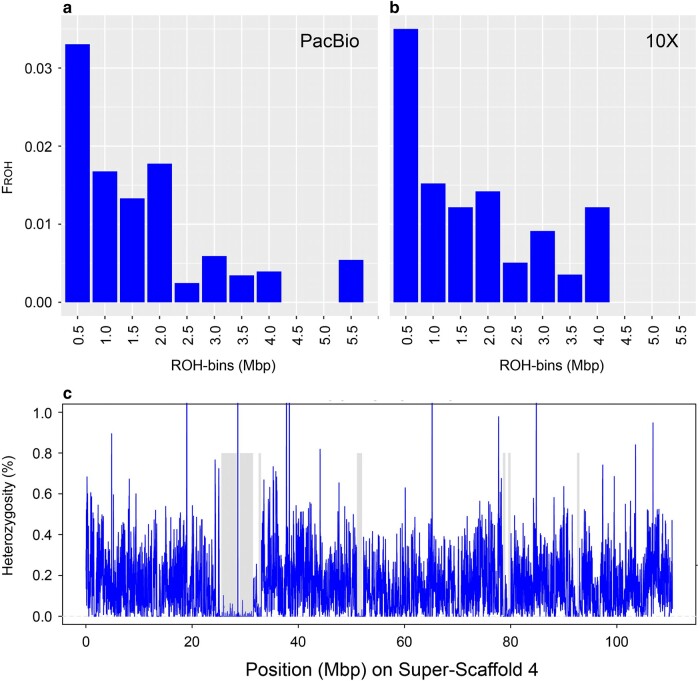
Fig. 8.
Comparison of inbreeding factors (FROH) based on the genome coverage of ROH between (a) the long-read assembly and (b) the linked-short-read assembly. ROH were identified with DARWINDOW, using a sliding-window-based approach, and sorted into respective length-bins. In both assemblies, 108 ROH over 500 kb were found; however, they appear to be more continuous in the long-read assembly as indicated by the longest ROH located on Super-Scaffold 4 (chromosome 8) of the respective assembly. A visual representation of this ROH is given in (c) and depicts the heterozygosity distribution of 20 kb windows over the scaffold in blue while identified ROH were marked as gray bars.