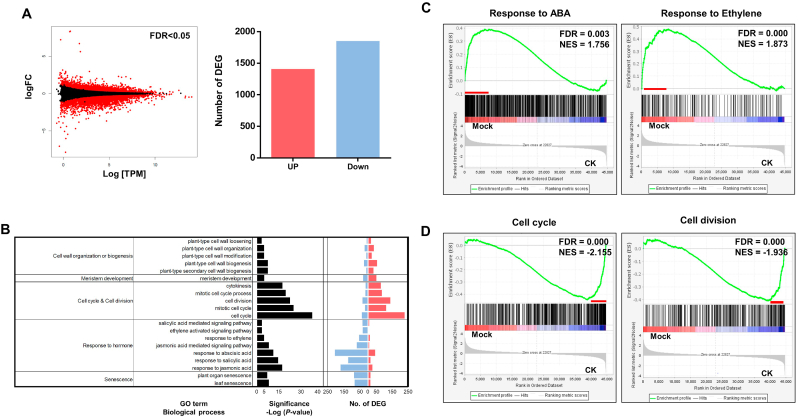
Fig. 3.
Transcriptome profiling of P. ginseng roots treated with or without CK. (A) MA plot shows differential expression between mock-and CK (Kinetin)-treated P. ginseng (Yeonpung) root samples. Red dots and bar graphs indicate the either up-(1,409) and down-(1,884) regulated genes with q < 0.05 and ≥ |1.5|-fold change. (B) Gene ontology (GO) enrichment analysis of differentially expressed genes (DEGs) identified by comparison of control and CK (Kinetin)-treated P. ginseng (Yeonpung) root samples. GO terms in biological process, with EASE score < 0.01 were selected (left panel). The number of up-regulated genes (red) and down-regulated genes (blue) categorized under the enriched GO terms are shown in the right panel. Enrichment plots for (C) response to ABA (GO:0010427, FDR = 0.0031986109, NES = 1.7566888) and ethylene-activated signaling pathway (GO:0009873, FDR = 0.00061904767, NES = 1.8739792), (D) cell cycle (GO:0007049, FDR = 0.000, NES = −2.1552036) and cell division (GO:0051301, FDR = 0.000, NES = −1.9369985).