Abstract
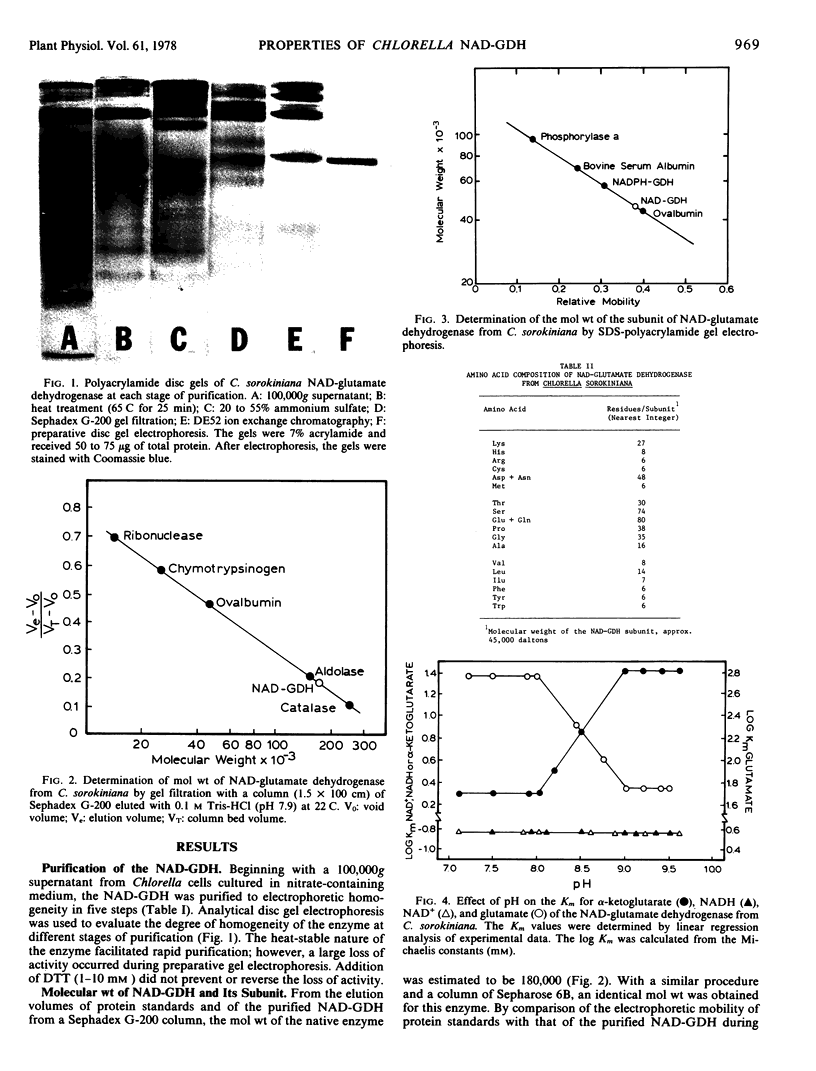
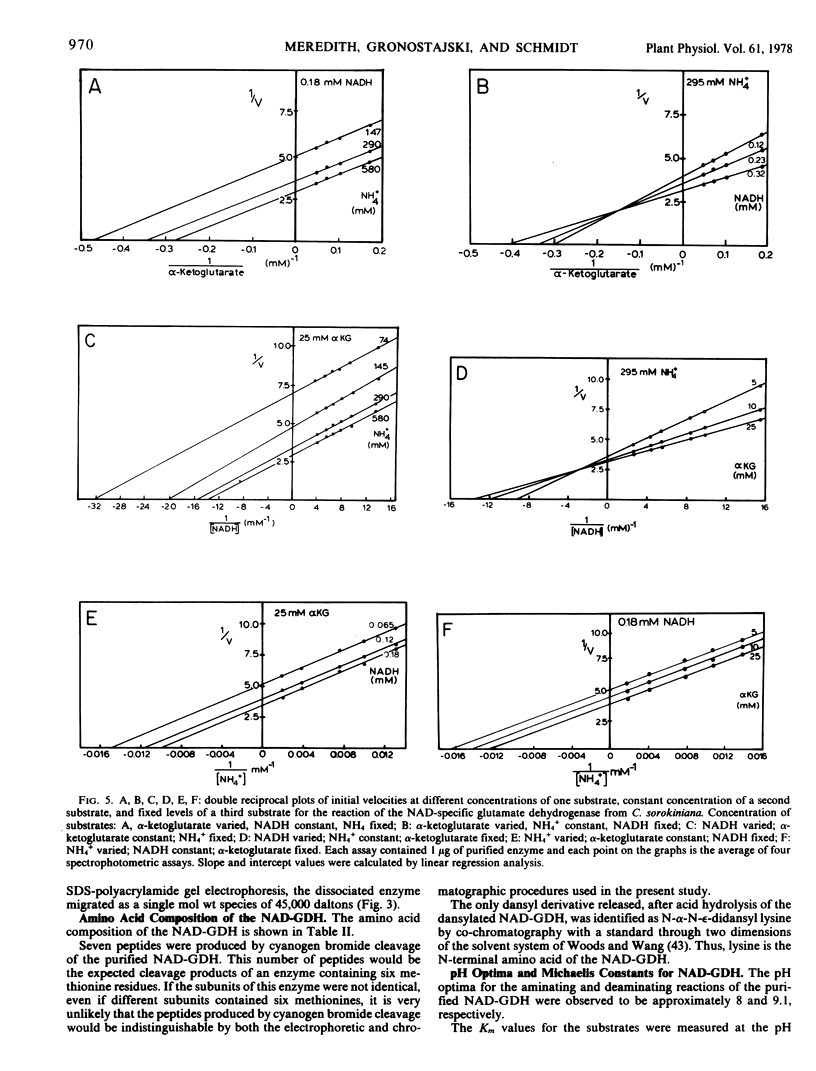
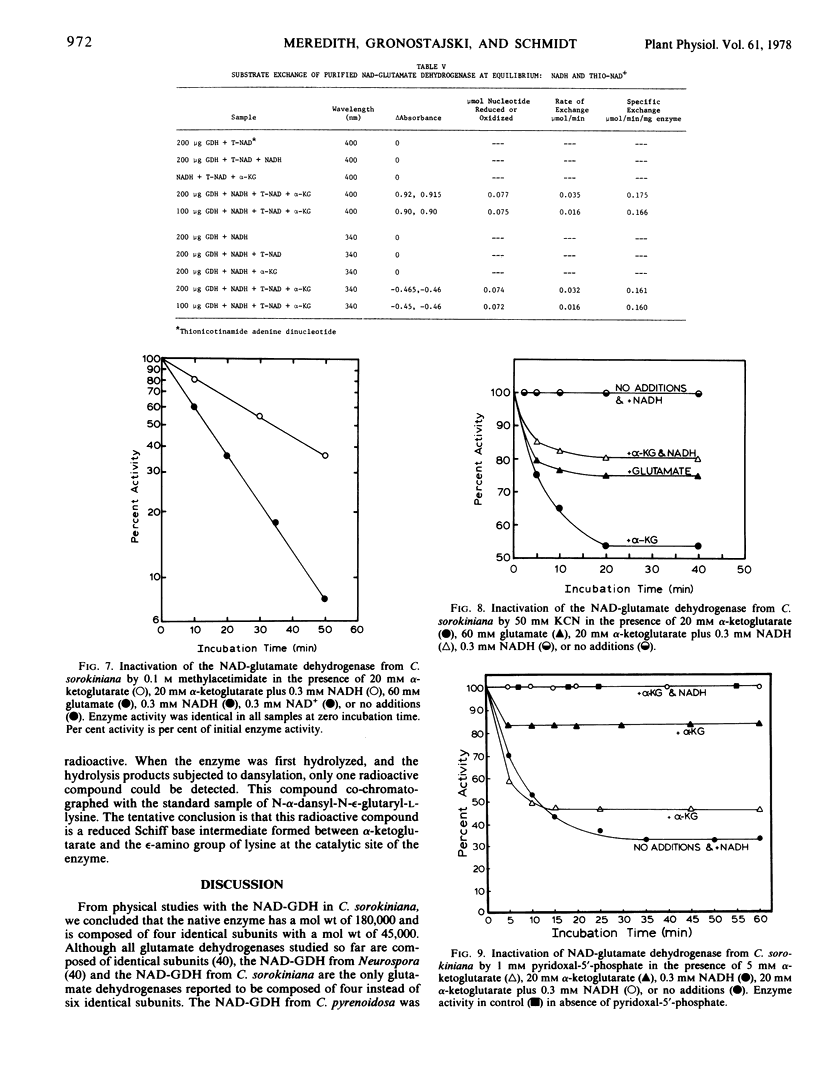
The nicotinamide adenine dinucleotide-specific glutamate dehydrogenase (l-glutamate:NAD+ oxidoreductase, EC 1.4.1.2) of Chlorella sorokiniana was purified 1,000-fold to electrophoretic homogeneity. The native enzyme was shown to have a molecular weight of 180,000 and to be composed of four identical subunits with a molecular weight of 45,000. The N-terminal amino acid was determined to be lysine. The pH optima for the aminating and deaminating reactions were approximately 8 and 9, respectively. The Km values for α-ketoglutarate, NADH, NH4+, NAD+, and l-glutamate were 2 mm, 0.15 mm, 40 mm, 0.15 mm, and 60 mm, respectively. Whereas the Km for α-ketoglutarate and l-glutamate increased 10-fold, 1 pH unit above or below the pH optima for the aminating or deaminating reactions, respectively, the Km values for NADH and NAD+ were independent of change in pH from 7 to 9.6. By initial velocity, product inhibition, and equilibrium substrate exchange studies, the kinetic mechanism of enzyme was shown to be consistent with a bi uni uni uni ping-pong addition sequence. Although this kinetic mechanism differs from that reported for any other glutamate dehydrogenase, the chemical mechanism still appears to involve the formation of a Schiff base between α-ketoglutarate and an ε-amino group of a lysine residue in the enzyme. The physical, chemical, and kinetic properties of this enzyme differ greatly from those reported for the NH4+-inducible glutamate dehydrogenase in this organism.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson B. M., Anderson C. D., Churchich J. E. Inhibition of glutamic dehydrogenase by pyridoxal 5'-phosphate. Biochemistry. 1966 Sep;5(9):2893–2900. doi: 10.1021/bi00873a017. [DOI] [PubMed] [Google Scholar]
- Andrews P. Estimation of the molecular weights of proteins by Sephadex gel-filtration. Biochem J. 1964 May;91(2):222–233. doi: 10.1042/bj0910222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Tana J., Cleland W. W. Rabbit muscle phosphofructokinase. I. Anomeric specificity; initial velocity kinetics. J Biol Chem. 1974 Feb 25;249(4):1263–1270. [PubMed] [Google Scholar]
- Bar-Tana J., Cleland W. W. Rabbit muscle phosphofructokinase. II. Product and dead end inhibition. J Biol Chem. 1974 Feb 25;249(4):1271–1276. [PubMed] [Google Scholar]
- Barash I., Sadon T., Mor H. Induction of a specific isoenzyme of glutamate dehydrogenase by ammonia in oat leaves. Nat New Biol. 1973 Aug 1;244(135):150–152. doi: 10.1038/newbio244150a0. [DOI] [PubMed] [Google Scholar]
- Baumann P., Wright B. E. The phosphofructokinase of Dictyostelium discoideum. Biochemistry. 1968 Oct;7(10):3653–3661. doi: 10.1021/bi00850a044. [DOI] [PubMed] [Google Scholar]
- Bigelow C. C. On the average hydrophobicity of proteins and the relation between it and protein structure. J Theor Biol. 1967 Aug;16(2):187–211. doi: 10.1016/0022-5193(67)90004-5. [DOI] [PubMed] [Google Scholar]
- Brown C. M., Burn V. J., Johnson B. Presence of glutamate synthase in fission yeasts and its possible role in ammonia assimilation. Nat New Biol. 1973 Nov 28;246(152):115–116. doi: 10.1038/newbio246115a0. [DOI] [PubMed] [Google Scholar]
- Cash D. J., Wilson I. B. The cyanide adduct of the aldolase dihydroxyacetone phosphate imine. J Biol Chem. 1966 Sep 25;241(18):4290–4292. [PubMed] [Google Scholar]
- DAVIS B. J. DISC ELECTROPHORESIS. II. METHOD AND APPLICATION TO HUMAN SERUM PROTEINS. Ann N Y Acad Sci. 1964 Dec 28;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- Gayler K. R., Morgan W. R. An NADP-dependent Glutamate Dehydrogenase in Chloroplasts from the Marine Green Alga Caulerpa simpliciuscula. Plant Physiol. 1976 Sep;58(3):283–287. doi: 10.1104/pp.58.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold M. H., Segel I. H. Neurospora crassa protein kinase. Purification, properties, and kinetic mechanism. J Biol Chem. 1974 Apr 25;249(8):2417–2423. [PubMed] [Google Scholar]
- Gore M. G., Greenwood C. Studies on the binary and ternary complexes formed by a Neurospora glutamate dehydrogenase and its substrates. Biochem Biophys Res Commun. 1975 Feb 17;62(4):997–1002. doi: 10.1016/0006-291x(75)90421-0. [DOI] [PubMed] [Google Scholar]
- Hodgins D. S., Abeles R. H. Studies of the mechanism of action of D-proline reductase: the presence on covalently bound pyruvate and its role in the catalytic process. Arch Biochem Biophys. 1969 Mar;130(1):274–285. doi: 10.1016/0003-9861(69)90034-4. [DOI] [PubMed] [Google Scholar]
- Hucho F., Markau U., Sund H. Studies of glutamate dehydrogenase. Characterization of histidine residues involved in the activity and association. Photoactivated labelling with pyridoxal 5'-phosphate. Eur J Biochem. 1973 Jan 3;32(1):69–75. doi: 10.1111/j.1432-1033.1973.tb02580.x. [DOI] [PubMed] [Google Scholar]
- Israel D. W., Gronostajski R. M., Yeung A. T., Schmidt R. R. Regulation of accumulation and turnover of an inducible glutamate dehydrogenase in synchronous cultures of Chlorella. J Bacteriol. 1977 May;130(2):793–804. doi: 10.1128/jb.130.2.793-804.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lea P. J., Miflin B. J. The occurrence of glutamate synthase in algae. Biochem Biophys Res Commun. 1975 Jan 2;64(3):856–862. doi: 10.1016/0006-291x(75)90126-6. [DOI] [PubMed] [Google Scholar]
- Leech R. M., Kirk P. R. An NADP-dependent L-glutamate dehydrogenase from chloroplasts of Vicia faba L. Biochem Biophys Res Commun. 1968 Aug 21;32(4):685–690. doi: 10.1016/0006-291x(68)90293-3. [DOI] [PubMed] [Google Scholar]
- LéJohn H. B., Jackson S. Allosteric interactions of a regulatory nicotinamide adenine dinucleotide-specific glutamate dehydrogenase from Blastocladiella. A molecular model for the enzyme. J Biol Chem. 1968 Jun 25;243(12):3447–3457. [PubMed] [Google Scholar]
- Miller R. E., Stadtman E. R. Glutamate synthase from Escherichia coli. An iron-sulfide flavoprotein. J Biol Chem. 1972 Nov 25;247(22):7407–7419. [PubMed] [Google Scholar]
- Pahlich E., Joy K. W. Glutamate dehydrogenase from pea roots: purification and properties of the enzyme. Can J Biochem. 1971 Jan;49(1):127–138. doi: 10.1139/o71-018. [DOI] [PubMed] [Google Scholar]
- Rasched I., Jörnvall H., Sund H. Studies of glutamate dehydrogenase. Identification of an amino group involved in the substrate binding. Eur J Biochem. 1974 Feb 1;41(3):603–606. doi: 10.1111/j.1432-1033.1974.tb03302.x. [DOI] [PubMed] [Google Scholar]
- SILVERSTEIN E., BOYER P. D. EQUILIBRIUM REACTION RATES AND THE MECHANISMS OF BOVINE HEART AND RABBIT MUSCLE LACTATE DEHYDROGENASES. J Biol Chem. 1964 Nov;239:3901–3907. [PubMed] [Google Scholar]
- Schmidt R. R. Continuous dilution culture system for studies on gene-enzyme regulation in synchronous cultures of plant cells. In Vitro. 1974 Nov-Dec;10(5-6):306–320. doi: 10.1007/BF02615312. [DOI] [PubMed] [Google Scholar]
- Shatilov V. R., Kretovich W. L. Glutamate dehydrogenases from Chlorella: forms, regulation and properties. Mol Cell Biochem. 1977 May 3;15(3):201–212. doi: 10.1007/BF01734109. [DOI] [PubMed] [Google Scholar]
- Talley D. J., White L. H., Schmidt R. R. Evidence for NADH- and NADPH-specific isozymes of glutamate dehydrogenase and the continuous inducibility of the NADPH-specific isozyme throughout the cell cycle of the eucaryote Chlorella. J Biol Chem. 1972 Dec 25;247(24):7927–7935. [PubMed] [Google Scholar]
- Weber K., Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969 Aug 25;244(16):4406–4412. [PubMed] [Google Scholar]
- Woods K. R., Wang K. T. Separation of dansyl-amino acids by polyamide layer chromatography. Biochim Biophys Acta. 1967 Feb 21;133(2):369–370. doi: 10.1016/0005-2795(67)90078-5. [DOI] [PubMed] [Google Scholar]



