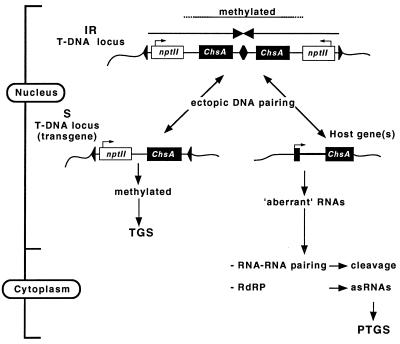
FIG. 6.
Proposed model for events that are initiated by an IR T-DNA locus and that lead to the posttranscriptional silencing of the host Chs genes or the transcriptional silencing of genes in a monomeric T-DNA locus (S). When two T-DNAs integrate into the genome as an IR, the sequences proximal to the center are preferentially methylated. If the transgene contains a promoter, this methylation is associated with TGS. A single-copy T-DNA locus (S) is methylated by the IR, and the pattern of methylation will be very similar to that of the IR. This may occur via ectopic DNA-DNA pairing between identical sequences of the IR and the S loci (left). An IR locus may also interact by DNA-DNA pairing with the homologous host gene(s) (right). This interaction does not lead to a dramatic change in DNA methylation but may cause a change in chromatin, which impedes regular transcription of the gene or RNA processing to some degree. In this way, “aberrant” or “unfinished” RNAs are produced, which either may be used as templates for the RNA-dependent RNA polymerase (17, 34) by which antisense RNAs are produced or may initiate a cycle of RNA-RNA pairing events by which they act as a kind of catalytic RNA (41). Both pathways result in the degradation of homologous transcripts and silence the genes posttranscriptionally.