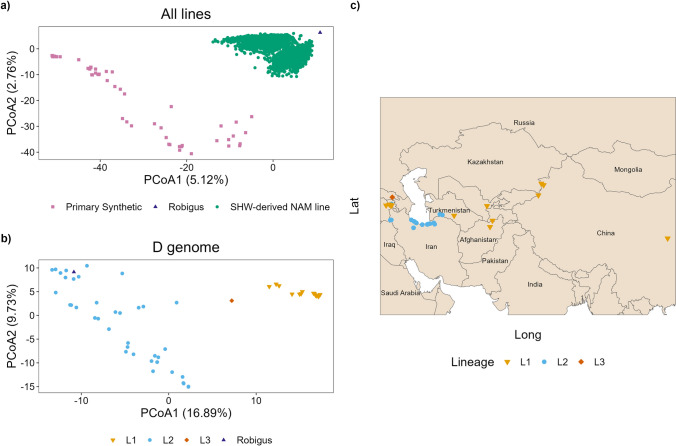
Fig. 2.
The NIAB_WW_SHW_NAM population is based on genetically diverse founders originating from a wide geographical area. Principal coordinate analysis (PCoA) of the first two principal coordinates for the NIAB_WW_SHW_NAM population genotypes and their parents (a) and the D sub-genome genetic markers for the primary synthetic hexaploid wheat donors and ‘Robigus’ (b), split into the three Ae. tauschii lineages (L1, L2, L3). The lineages of Ae. tauschii were assigned to the distinct clusters using overlapping genotypes between the present study and Gaurav et al. (2022). c A map showing the passport data location of 43 Ae. tauschii (diploid, D genome) accessions captured in the primary SHW (hexaploid, ABD genome)