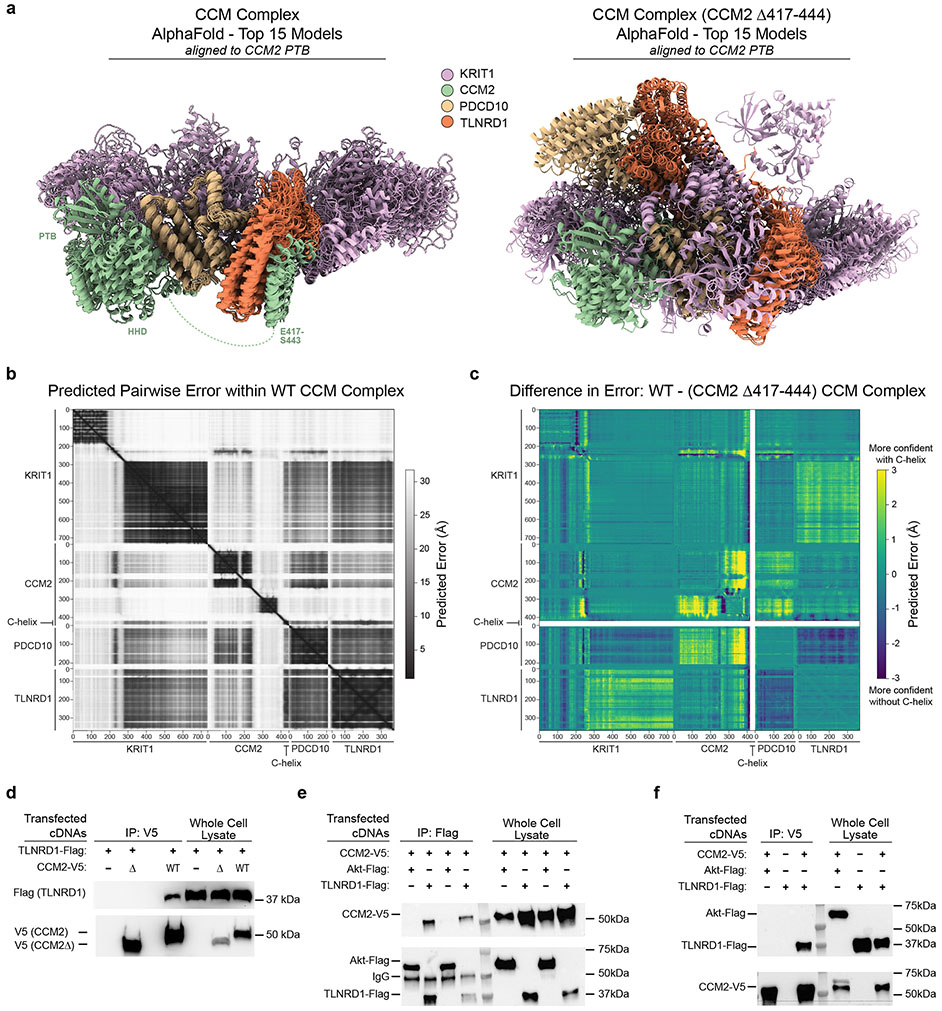
Extended Data Fig. 9. TLNRD1 interaction with the CCM complex depends on the CCM2 C-terminal helix.
a. The 15 top-ranking AlphaFold2 models of the CCM complex are shown, as predicted with and without the presence of CCM2 residues 417-444, and aligned to the CCM2 PTB domain (residues 55-237). The complex is consistently predicted in a high-confidence arrangement (left panel), with most variability in positions of the HHD domain of CCM2 and flexible regions of Krit1. Interactions are predicted between all members of the complex, including published interactions between CCM2-PDCD10. In contrast, multiple conformations are predicted in the absence of the CCM2 C-terminal helix (right panel).
b. Predicted Alignment Error (PAE, Å) of all pairwise residue combinations in the WT CCM complex (extracted and plotted using AlphaPickle 82); lower error indicates higher confidence.
c. Differences in PAE (Å) between the full CCM complex and the CCM complex lacking the CCM2 C-terminal helix; larger numbers represent higher confidence in the presence of the helix. The thick white lines correspond to deleted helix residues, which are omitted from comparison. Predictions within individual domains and proteins are largely unaffected, but within CCM2, the HHD and subsequent loops are predicted with reduced confidence upon helix deletion (yellow). Between domains and proteins, the largest differences are reduced confidence interactions between CCM2 and PDCD10 and between Krit1 and TLNRD1. Summing over the entire matrices, the increase in predicted error with deletion of the helix is 0.7Å (22.9 vs 23.6 Å, p=10−56, two-tailed t-test).
d. FLAG-tagged TLNRD1 and/or V5-tagged CCM2 full length (“WT”) or C-terminal truncation (“Δ”) were expressed in HEK293T cells, as indicated. Extracts were co-immunoprecipitated with rabbit anti-V5 and blotted with mouse anti-Flag (top) or mouse anti-V5 (bottom). For gel source data, see Supplementary Figure 1b. Similar results were seen in 2 separate experiments.
e. HEK293 cells were transfected with V5-tagged CCM2 and either Flag-tagged TLNRD1 or Flag-tagged Akt (negative control). Cell lysates were either immunoprecipitated with mouse anti-Flag antibody-bound beads (IP Flag), or loaded directly on the gel (Input). The membranes were first probed with rabbit anti-V5 to detect CCM2 in the Flag precipitant and confirm the transfection of CCM2-V5. The membranes were then re-blotted with rabbit anti-TLNRD1 to evaluate the efficiency of Flag immunoprecipitation and validate the transfection of Akt-Flag and TLNRD1-Flag. Each pair of lanes came from independent biological replicates. For gel source data, see Supplementary Figure 1c. Similar results were seen in 2 separate experiments.
f. HEK293 cells were transfected with V5-tagged CCM2 and Flag-tagged TLNRD1, or, as negative controls, either with CCM2-V5 and Akt-Flag or only TLNRD1-Flag. Cell lysates were either immunoprecipitated with anti-V5 beads (IP V5), or loaded directly on the gel (Input). The membranes were first blotted for Flag to detect TLNRD1 in the V5 precipitant and validate the transfection of Akt-Flag and TLNRD1-Flag. The membranes were re-blotted for V5 to evaluate the efficiency of V5 immunoprecipitation and confirm the transfection of CCM2-V5. For gel source data, see Supplementary Figure 1d. Similar results were seen in 2 separate experiments.