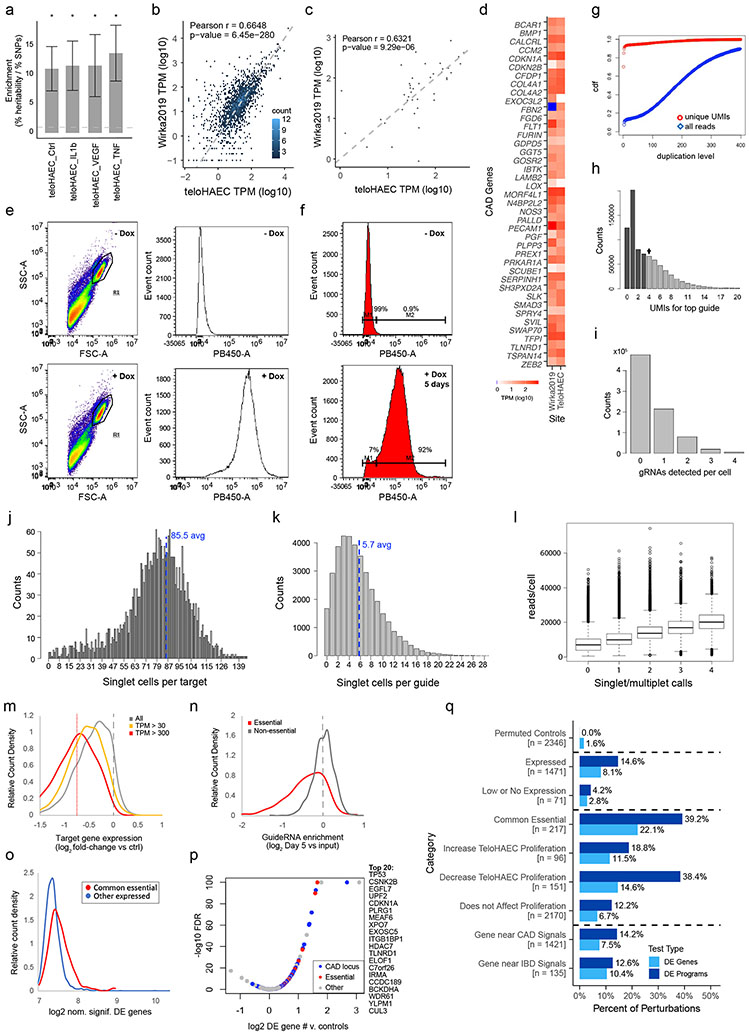
Extended Data Fig. 1. Establishing the TeloHAEC CRISPRi model and Perturb-seq details.
a. Enrichment of CAD heritability in TeloHAEC enhancers, from Stratified Linkage Disequilibrium Score Regression analysis (S-LDSC, see Methods), where enrichment is the percentage of heritability explained by variants in enhancers (%heritability), divided by the percentage of variants in enhancers (%SNPs). Enhancers in TeloHAEC (treated under the indicated conditions) were identified from ATAC-seq and H3K27ac ChIP-seq data (n=6 for control ATAC, 3 for IL-1β, TNFα or VEGF ATAC, 4 for control ChIP, and 2 for IL-1β, TNFα or VEGF ChIP) by the Activity-by-Contact model. Error bars: standard error around the enrichment estimate, calculated by S-LDSC using jackknife (which resamples the data used for calculating heritability enrichment). P.values were calculated using the S-LDSC method28, and FDR by the Benjamini-Hochberg method. *: FDR<0.05, with specific FDR values of: Ctrl; 0.037, IL-1β; 0.015, TNFα; 0.020 and VEGF; 0.041. Full S-LDSC results can be found in Supplementary Table 27.
b. Scatter density plot of human right coronary artery endothelial cell single cell RNA-seq pseudobulk gene expression (from 69) versus teloHAEC pseudobulk gene expression, for genes perturbed in this study. Among the perturbed genes in teloHAEC, 2,107 genes are expressed at TPM > 1 in healthy or diseased RCAECs. R and p.values from two sided Pearson correlation test.
c. Scatter plot of the 41 V2G2P genes, comparing single cell RNA-seq pseudobulk expression (in TPM) in human right coronary artery endothelial cells to TeloHAEC. R and p.values from two sided Pearson correlation test.
d. Heatmap of gene expression (log10 TPM) of the 41 V2G2P genes in diseased right coronary artery ECs and in teloHAEC. 40 out of 41 CAD associated genes are expressed at >1 TPM in RCAECs. FBN2 is lowly expressed in the human right coronary artery endothelial cells.
e. FACS showing dox inducibility of KRAB-dCas9-IRES-BFP in TeloHAEC, after sorting but before the screen. Left panels: gating for viable individual cells. Right panels: Counts of gated cells by fluorescence intensity in the BFP/PB450 channel.
f. BFP channel counts of cells grown in parallel and concurrently with cells for the Perturb-seq screen. After expansion to 120M cells, transduction, selection and 5-day doxycycline treatment, 92% of cells remain BFP positive.
g. Cumulative distribution fraction for duplication levels of unique CBC-UMI-Guide combinations in deeply-sequenced dialout libraries (“unique UMIs”, red) or all guide reads (blue) versus duplication level. Requiring 4 duplicates (dotted line) eliminates 90% of CBC-UMI-guide combinations (likely PCR chimeras), while retaining >85% of total guide reads.
h. UMIs for top guide per CBC. Arrow: the chosen 4 UMI threshold.
i. Counts of singlets (1 gRNA, black bar), doublets (2) and higher multimers, as well as cells with no guide called (0), at the chosen thresholds of 4 UMIs for the top guide and 4 or more fold fewer for the next most frequent guide.
j. Histogram of counts of singlet cells per target. Dotted line: average.
k. Histogram of counts of singlet cells per guide. Dotted line: average.
l. Read UMI counts for all transcripts per cell by singlet/multiplet status. Median UMIs per singlet cell was 9,997, and average was 10,870. The median for cells with no guide called was 7,125, indicating that low guide UMI count is associated with low overall UMI count. Median UMIs for doublets was 13,723, 37.3% more than singlets. Assuming that droplets with two cells will have double the number of reads, this suggests 37% of doublets are due to two cells (9.3% of cells with guides) while the remainder (15.7% of cells with guides) are due to two guides in one cell, very close to the expectation from the infection MOI of 15%. n=352686, 214449, 79744, 19195 and 5345 cells with 0, 1, 2, 3, or 4 guides, respectively. Boxplot center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; points, outliers.
m. Distribution of knockdown efficiency across target genes (log2 expression in cells containing guideRNAs targeting the gene versus in cells containing negative control guideRNAs). Gray line: all targeted genes. Yellow and red lines: Genes expressed at >30 and >300 TPM, respectively. Red dotted vertical line: 40% knockdown (average for 300+ TPM target genes).
n. Distribution of fitness effects across all guideRNAs (log2 ratio of guide frequency in singlet cells from the Perturb-seq experiment after 5 days of CRISPRi induction compared to guide frequency in the original guideRNA library). Guides targeting common essential genes (red) were depleted more frequently than guideRNAs targeting other genes.
o. Number of nominally significant differentially expressed (DE) genes per perturbed target (genes with raw p <0.01, and fold change >1.15 from EdgeR DE analysis). Perturbations that affected the transcriptome were those that significantly increased the number of nominally significant DE genes relative to the 48 targeted negative control genes (not expressed in TeloHAEC). Dotted line: 95th percentile number of DE genes for negative controls. 245 perturbations had a significant effect on the transcriptome, FDR <0.05 (10.7% of all targets that were not negative controls: including 31.9% of common essential genes (red, as per panel n), and 9.0% of other genes (blue)).
p. Volcano plot showing log2 (# DE genes for target)/(avg. # DE genes for non-expressed controls) versus −log10 FDR (capped at 100). Right: Symbols for target genes with the strongest effects.
q. Percent of perturbations that have a significant transcriptional effect in Perturb-seq, as defined by either (i) “DE Genes”: perturbations with significant effect on the transcriptome, as compared to 48 non-expressed negative control promoters, by binomial test (see Methods) or (ii) “DE Programs”: perturbations that lead to significant changes in program expression by MAST with 10X lane correction (FDR < 0.05).
Permuted Controls: Simulated negative controls, where statistical tests were performed on randomly drawn cells that carry negative control or safe-targeting guides.
Expressed: Genes with >1 transcripts per million (TPM) in teloHAEC control bulk RNA-seq.
Low or No Expression: Genes with less than or equal to 1 TPM.
Common Essential: Common essential genes from DepMap147.
TeloHAEC Proliferation: Fitness effects observed in the Perturb-seq experiment, by comparing guide frequencies (see Methods). Increase: >15% increase in guide frequency (FDR < 0.05), Decrease: >15% decrease in guide frequency (FDR < 0.05).
Gene near CAD GWAS signals: Expressed genes nearby any CAD GWAS signal (2 closest on each side, and all within +/−500kb).
Gene near IBD signals: Expressed genes nearby 10 selected IBD GWAS signals (closest 2 genes & all within +/−500kb), with no genes overlapping those for CAD signals.