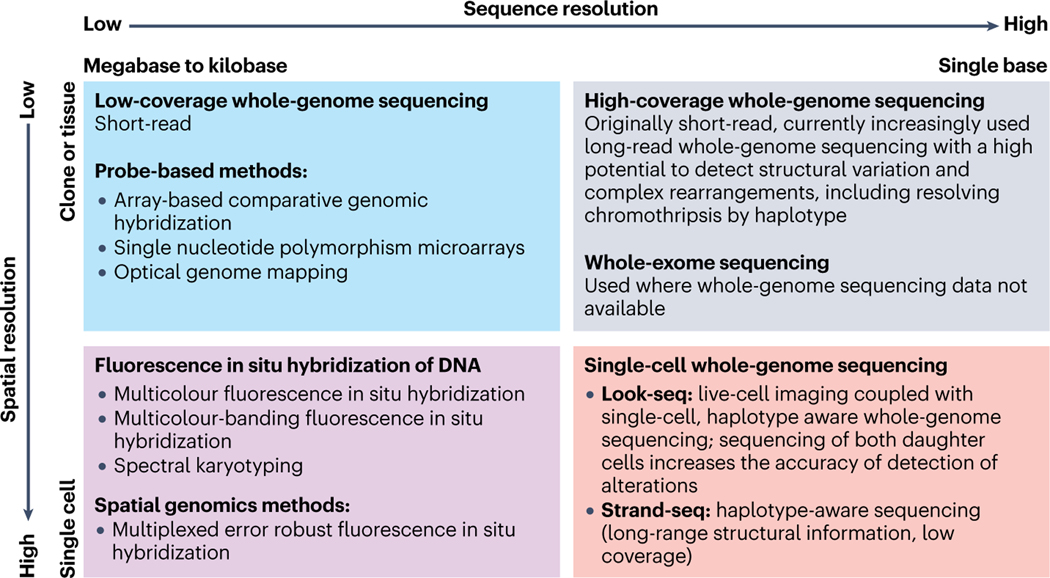
Fig. 1. Methods to detect chromothripsis with different spatial and sequence resolution.
Complementary techniques applied for the detection of chromothripsis vary in the precision of sequence resolution (ranging from the megabases to single nucleotides, horizontal axis) and spatial resolution (from clones or tissue samples to single cells, vertical axis). Top left square: examples of high-throughput techniques with relatively low spatial/sequence resolution: low coverage whole genome sequencing (WGS) and in situ hybridization probe-based methods; top right square: the highest sequence resolution with low spatial resolution is achieved by high coverage WGS (might be combined with whole exome sequencing); bottom left square: advanced fluorescent-in-situ-hybridization techniques provide the highest (single-cell) spatial/relatively low (within kilobases to megabases) sequence resolution; bottom right square: single-cell WGS aims at achieving the highest spatial/sequence resolution, but the scarcity of genomic material remains a significant challenge.